Table of Contents
Welcome to the SoilWise Technical Documentation!
SoilWise Technical Documentation currently consists of the following sections:
- Technical Components
- Interfaces
- Infrastructure
- Glossary
- Printable version - where you find all sections composed in one page, that can be easily printed using Web Browser options
Essential Terminology
A full list of terms used within this Technical Documentation can be found in the Glossary. The most essential ones are defined as follows:
- (Descriptive) metadata: Summary information describing digital objects such as datasets and knowledge resources.
-
Metadata record: An entry in e.g. a catalogue or abstracting and indexing service with summary information about a digital object.
-
Data: A collection of discrete or continuous values that convey information, describing the quantity, quality, fact, statistics, other basic units of meaning, or simply sequences of symbols that may be further interpreted formally (Wikipedia).
-
Dataset: (Also: Data set) A collection of data (Wikipedia).
-
Knowledge: Facts, information, and skills acquired through experience or education; the theoretical or practical understanding of a subject. SoilWise mainly considers explicit knowledge -- Information that is easily articulated, codified, stored, and accessed. E.g. via books, web sites, or databases. It does not include implicit knowledge (information transferable via skills) nor tacit knowledge (gained via personal experiences and individual contexts). Explicit knowledge can be further divided into semantic and structural knowledge:
- Semantic knowledge: Also known as declarative knowledge, refers to knowledge about facts, meanings, concepts, and relationships. It is the understanding of the world around us, conveyed through language. Semantic knowledge answers the "What?" question about facts and concepts.
- Structural knowledge: Knowledge about the organisation and interrelationships among pieces of information. It is about understanding how different pieces of information are interconnected. Structural knowledge explains the "How?" and "Why?" regarding the organisation and relationships among facts and concepts.
- Knowledge resource: A digital object, such as a document, a web page, or a database, that holds relevant explicit knowledge.
Release notes
| Date | Action |
|---|---|
| 27. 2. 2025 | v2.1 Released: For D2.2 Developed & Integrated DM components, v2 D3.2 Developed & Integrated KM components, v2 and D4.2 Repository infrastructure, components and APIs, v2 purposes |
| 26. 2. 2025 | Link liveliness assessment tool updated |
| 25. 2. 2025 | Metadata Validation updated |
| 20. 2. 2025 | Knowledge Graph component updated |
| 19. 2. 2025 | Apache Solr component added |
| 19. 2. 2025 | Storage updated |
| 19. 2. 2025 | Catalogue updated |
| 14. 2. 2025 | Metadata Validation updated |
| 13. 2. 2025 | Metadata Augmentation updated |
| 7. 2. 2025 | Interfaces description updated |
| 30. 9. 2024 | v2.0 Released: For D2.1 Developed & Integrated DM components, v1 D3.1 Developed & Integrated KM components, v1 and D4.1 Repository infrastructure, components and APIs, v1 purposes |
| 30. 9. 2024 | Technical Components functionality updated according to first SoilWise repository prototype |
| 27. 8. 2024 | APIs section restructured |
| 20. 8. 2024 | Knowledge Graph component added |
| 13. 8. 2024 | Metadata Authoring component added |
| 1. 7. 2024 | Metadata Augmentation component added |
| 30. 4. 2024 | v1.0 Released: For D1.3 Architecture Repository v1 purposes |
| 27. 3. 2024 | Technical Components restructured according to the architecture from Brugges Technical Meeting |
| 27. 3. 2024 | v0.1 Released: Technical documentation based on the Consolidated architecture |
| 10. 2. 2024 | Technical Documentation was initialized |
Technical Components
Introduction
The SoilWise Catalogue (SWC) architecture aims towards efficient facilitation of soil data & knowledge management. It seamlessly gathers, processes, and disseminates data from diverse sources. The system prioritizes high-quality data dissemination, knowledge extraction and interoperability while user management and monitoring tools ensure secure access and system health. Note that, SWC primarily serves to power Decision Support Systems (DSS) rather than being a DSS itself.
The presented architecture represents an outlook and a framework for ongoing SoilWise development. As such, the implementation has been following intrinsic (within the SoilWise project) and extrinsic (e.g. EUSO development Mission Soil Projects) opportunities and limitations. The presented architecture corresponds to the current state of third SoilWise prototype delivery. Modifications during the implementation will be incorporated into the final version of the SoilWise architecture due M42.
This section lists technical components for building the SoilWise Catalogue as forseen in the architecture design:
- Harvester
- Repository Storage
- Metadata Catalogue
- Soil Companion (Chatbot)
- Data publication support
- Metadata Validation
- Metadata Augmentation
- Knowledge Graph
- Data & Knowledge Administration Console
- User Management and Access Control
- System & Usage Monitoring
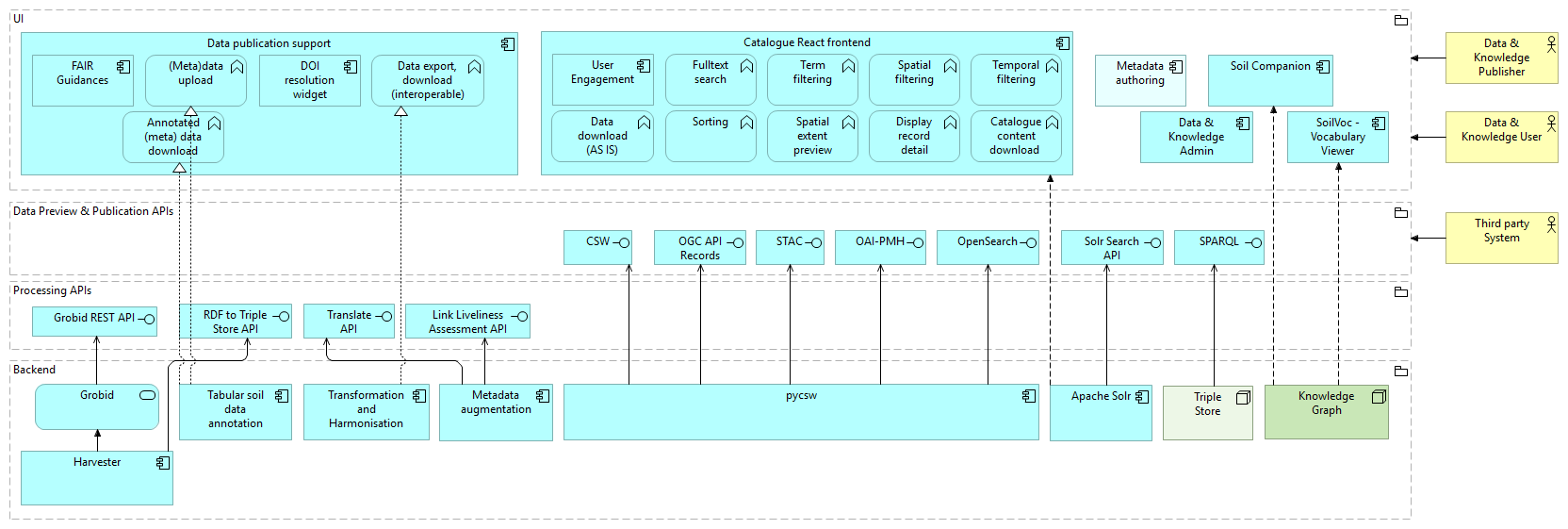
 Fig. 1: A high-level overview of SoilWise Repository architecture
Fig. 1: A high-level overview of SoilWise Repository architecture
A full version of architecture diagram is available at: https://soilwise-he.github.io/soilwise-architecture/.
Harvester
Info
Current version: 0.2.0
Technology: Git pipelines
Release: https://doi.org/10.5281/zenodo.14923563
Project: Harvesters
The Harvester component is dedicated to automatically harvest sources to populate SWR with metadata on datasets and knowledge sources.
Introduction
Overview and Scope
Metadata harvesting is the process of ingesting metadata, i.e. evidence on data and knowledge, from remote sources and storing it locally in the catalogue for fast searching. It is a scheduled process, so local copy and remote metadata are kept aligned. Various components exist which are able to harvest metadata from various (standardised) API's. SoilWise aims to use existing components where available.
The harvesting mechanism relies on the concept of a universally unique identifier (UUID) or unique resource identifier (URI) that is being assigned commonly by metadata creator or publisher. Another important concept behind the harvesting is the last change date. Every time a metadata record is changed, the last change date is updated. Just storing this parameter and comparing it with a new one allows any system to find out if the metadata record has been modified since last update. An exception is if metadata is removed remotely. SoilWise Repository can only derive that fact by harvesting the full remote content. Discussion is needed to understand if SWR should keep a copy of the remote source anyway, for archiving purposes. All metadata with an update date newer then last-identified successfull harvester run are extracted from remote location.
A harvesting task typically extracts records with update-date later then the last-identified successfull harvester run. In case the remote system supports such a filter, else the full set is harvested.
Local improvements to metadata records should be stored separately from the harvested content for the following reasons:
- The harvesting is periodic so any local change to harvested metadata will be lost during the next run.
- The change date may be used to keep track of changes so if the metadata gets changed, the harvesting mechanism may be compromised.
If inconsistencies with imported metadata are identified, we can add a statement to the graph of such inconsistencies. We can also notify the author of the inconsistency so they can fix the inconsistency on their side.
A governance aspect still under discussion is if harvested content is removed as soon as a harvester configuration is removed, or when records are removed from the remote endpoint. The risk of removing content is that relations within the graph are breached. An alternative is to indicate the record has been archived by the provider.
On top of a unique identification, SWR also captures a unique calculated string (a hash) for the harvested content. This allows to identify changes even if the update date has not changed.
Typical tasks of a harvester:
- Define a harvester job
- Schedule (on request, weekly, daily, hourly)
- Endpoint / Endpoint type (example.com/csw -> OGC:CSW)
- Apply a filter (only records with keyword='soil-mission')
- Understand success of a harvest job
- overview of harvested content (120 records)
- which runs failed, why? (today failed -> log, yesterday successfull -> log)
- Monitor running harvestors (20% done -> cancel)
- Define behaviours on harvested content
- skip records with low quality (if test xxx fails)
- mint identifier if missing ( https://example.com/data/{uuid} )
- a model transformation before ingestion ( example-transform.xsl / do-something.py )
Intended Audience
Harvester is a backend component, therefore we only expect a maintenance role:
- SWC Administrator monitoring the health status, logs... Administrators can manually start a specific harvesting pipelines.
Resource Types
Metadata for following resource types are foreseen to be harvested:
- Data & Knowledge Resources (datasets, services, software, documents, articles, videos)
- Organisations, Projects, LTE, Living labs initiatives
- News items from relevant websites
These entities relate to each other as:
flowchart LR
people -->|memberOf| o[organisations]
o -->|partnerIn| p[projects]
p -->|produce| d[data & knowledge resources]
o -->|publish| d
d -->|describedIn| c[catalogues]
p -->|part-of| fs[Fundingscheme]
Datasets
Metadata records of datasets are, for the first iteration, primarily imported from the ESDAC, INSPIRE GeoPortal, BonaRes, Cordis/OpenAire, ISRIC, FAO, and EEA. In later iterations SoilWise aims to include other projects and portals, such as national or thematic portals. These repositories contain large number of datasets. Selection of key datasets concerning the SoilWise scope is a subject of know-how to be developed within SoilWise.
Knowledge sources
Compared to datasets, knowledge sources are typically very heterogeneous (reports, articles, websites, video's) and collected in a variety of repositories. Soilwise endorses projects to use persistent repositories, with sufficient options for metadata capture. For the acadmic community for example, inclusion in OpenAire is a prerequisite to be included in SWR. This allows SWR to use the OpenAire functionalities to collect evidence about the resources.
The SoilWise project team is still exploring which knowledge resources to include. As an example, an important cluster of knowledge sources may be seen academic articles and report deliverables from Mission Soil Horizon Europe projects. These resources are accessible from Cordis and OpenAire, filteres by the grantnumber of the projects. Extracting content from Cordis and OpenAire can be achieved using a harvesting task (using the Cordis schema, extended with post processing). For the first iteration, SoilWise aims to achieve this goal. In future iterations new knowledge sources may become relevant, we will investigate at that moment what is the best approach to harvest them.
Projects and organisations
Project details are extracted from Cordis. Discussion is ongoing how to improve this process, for example to understand if projects should be included which do not have European funding.
Indivicuals and organisations are typically mentioned as contact, author or owner in metadata records, as well as participant or funder in projects.
A challenge for SWR is to understand the alignment between those individuals and organisations, to enable users to understand the relations between projects, organisations and resources.
News items
A need has been expressed to be informed about ongoing Soil Mission projects. For that reason a harvesting mechanism has been set up which extracts and aggregates from the various Soil Mission Project websites the news items published in their websites. A common protocol, RSS/Atom feeds, implemented by most of the project websites is used to extract that information. At the moment we are investigating if we can also extract anounced upcoming events, for example via the iCalendar protocol, but we already noticed that this protocol has vert little adoption.
Adoption of standards
With respect to harvesting, it is important to note that a wide range of levels of adoption of standards is implemented by repositories. Both for metadata models, identification, as well as access protocols. This will, in some cases, make it necessary to develop customized harvesting and metadata extraction processes. It also means that informed decisions need to be made on which resources to include, based on priority, required efforts and available capacity.
Key features
The Harvester component currently comprises of the following functions:
- Harvest records from metadata and knowledge resources
- Metadata Harmonization
- Metadata RDF Turtle Serialization
- RDF to Triple Store
- Duplication Identification
Harvest records from metadata and knowledge resources
Note, the second SoilWise Repository prototype contained 19,324 harvested metadata records (to date 14.2.2025).
CORDIS
European Research projects typically advertise their research outputs via Cordis. This makes Cordis a likely candidate to discover research outputs, such as reports, articles and datasets. Cordis does not capture many metadata properties. In those cases where a resource is identified by a DOI, additional metadata can be found in OpenAire via the DOI. The scope of projects, from which to include project deliverables is still under discussion.
Which projects to include is derived from 2 sources:
- ESDAC maintains a list of historic EU funded research projects
- Mission soil platform maintains a list of current Mission soil projects
A script fetches the content from these 2 sources and prepares relevant content for the CORDIS and OpenAire harvesting. The content in these pages is unstructured html. The content is scraped using a python library. This is not optimal, because the scraper expects a dedicated html structure, which is fragile.
Results of the scrape activity are stored in table harvest.projects. For each project a Record control number (RCN) is retrieved from the Cordis knowledge graph. This RCN could be used to filter OpenAire, however OpenAire can also be filtered using project grant number. At this moment in time the Cordis Knowledge graph does not contain the Mission Soil projects yet.
Currently we do not harvest resources from Cordis which do not have a DOI. This includes mainly progress reports of the projects.
OpenAire
For those resources, discovered via Cordis/ESDAC, and identified by a DOI, a harvester fetches additional metadata from OpenAire. OpenAire is a catalogue initiative which harvests metadata from popular scientific repositories, such as Zenodo, Dataverse, etc.
Not all DOI's registered in Cordis are available in OpenAire. OpenAire only lists resources with an open access license. Other DOI's can be fetched from the DOI registry directly or via Crossref.org. This work is still in preparation.
Records in OpenAire are stored in the Open Aire Research Graph (OAF) format, which is transformed to a metadata set based on Dublin Core.
OGC-CSW
Many (spatial) catalogues advertise their metadata via the catalogue Service for the Web standard, such as INSPIRE GeoPortal, Bonares, ISRIC. The OWSLib library is used to query records from CSW endpoints. A filter can be configured to retrieve subsets of the catalogue.
Incidentally, records advertised as CSW also include a DOI reference (Bonares/ISRIC). Additional metadata for these DOI's is extracted from OpenAire/Crossref.
INSPIRE
Although INSPIRE Geoportal does offer a CSW endpoint, due to a technical reasons, we have not been able to harvest from it. Instead we have developed a dedicated harvester via the Elastic Search API endpoint of the Geoportal. If at some point the technical issue has been resolved, use of the CSW harvest endpoint is favourable.
ESDAC
The ESDAC catalogue is an instance of Drupal CMS. We have developed a dedicated harvester to scrape html elements to extract Dublin Core metadata from ESDAC html elements. Metadata is extracted for datasets, maps (EUDASM) and documents. Incidentally a DOI is mentioned as part of the HTML, this DOI is then used as identifier for the resource, else the resource url is used as identifier. If the DOI is not known to the system yet, OpenAire will be queried to capture additional metadata on the resource.
Prepsoil portal
Prepsoil is build on a headless CMS. The CMS at times provides an API to retrieve datasets, knowledge items, living labs, lighthouses and communities of practice. The API provides minimal metadata, incidentally a DOI is included. DOI is used to capture additional metadata from OpenAire.
News feeds
From the project websites mentioned at https://mission-soil-platform.ec.europa.eu/project-hub/funded-projects-under-mission-soil a harvester algorythm fetches the contents of the RSS feed, if the website provides one. The harvested entries are stored on a database.
Metadata Harmonization
Once stored in the harvest sources database, a second process is triggered which harmonizes the sources to the desired metadata profile. These processes are split by design, to prevent that any failure in metadata processing would require to fetch remote content again.
Table below indicates the various source models supported
| source | platform |
|---|---|
| Dublin Core | Cordis |
| Extended Dublin core | ESDAC |
| Datacite | OpenAire, Zenodo, DOI |
| ISO19115:2005 | Bonares, INSPIRE |
Metadata is harmonised to a DCAT RDF representation.
For metadata harmonization some supporting modules are used, OWSlib is a module to parse various source metadata models, including iso19139:2007. A transformation script from semic-eu/iso19139-to-dcat-ap.xslt in combination with lxml and rdflib is used to convert iso19139:2007 metadata to RDF, serialised as turtle.
Harmonised metadata is either transformed to iso19139:2007 or Dublin Core and then ingested by the pycsw software, used to power the SoilWise Catalogue, using an automated process running at intervals. At this moment the pycsw catalogue software requires a dedicated database structure. This step converts the harmonised metadata database to that model. In next iterations we aim to remove this step and enable the catalogue to query the harmnised model directly.
Metadata Augmentation
The metadata augmentation processes are described elsewhere, what is relevant here is that the output of these processes is integrated in the harmonised metadata database.
Metadata RDF turtle serialization
The harmonised metadata model is based on the DCAT ontology. In this step the content of the database is written to RDF.
Harmonized metadata is transformed to RDF in preparation of being loaded into the triple store (see also Knowledge Graph).
RDF to Triple store
This is a component which on request can dump the content of the harmonised database as an RDF quad store. This service is requested at intervals by the triple store component. In a next iteration we aim to push the content to the triple store at intervals.
Duplication indentification
A resource can be described in multiple Catalogues, identified by a common identifier. Each of the harvested instances may contain duplicate, alternative or conflicting statements about the resource. SoilWise Repository aims to persist a copy of the harvested content (also to identify if the remote source has changed). For this iteration we store the first copy, and capture on what other platforms the record has been discovered. OpenAire already has a mechanism to indicate in which platforms a record has been discovered, this information is ingested as part of the harvest. An aim of this exercise is also to understand in which repositories a certain resource is advertised.
Visualization of source repositories is in the first development iteration available as a dedicated section in the SoilWise Catalogue.
Architecture
Technological Stack
| Technology | Description |
|---|---|
| Git actions/pipelines | Automated processes which run at intervals or events. Git platforms typically offer this functionality including extended logging, queueing, and manual job monitoring and interaction (start/stop). |
Main Sequence Diagram
Each harvester runs in a dedicated container. The result of the harvester is ingested into a (temporary) storage. Follow up processes (harmonization, augmentation, validation) pick up the results from the temporary storage.
flowchart LR
c[CI-CD] -->|task| q[/Queue\]
r[Runner] --> q
r -->|deploys| hc[Harvest container]
hc -->|harvests| db[(temporary storage)]
hc -->|data cleaning| db[(temporary storage)]
Integrations & Interfaces
The Automatic metadata harvesting component will show its full potential when being in the SWR tightly connected to (1) SWR Catalogue, (2) Metadata authoring and (3) ETS/ATS, i.e. test suites.
Key Architectural Decisions - Harvesting Strategy
OGC-CSW
Many (spatial) catalogues advertise their metadata via the catalogue Service for the Web standard, such as INSPIRE GeoPortal, Bonares, ISRIC.
CORDIS - OpenAire
Cordis does not capture many metadata properties. We harvest the title of a project publication and, if available, the DOI. In those cases where a resource is identified by a DOI, additional metadata can be found in OpenAire via the DOI. For those resources a harvester fetches additional metadata from OpenAire.
A second mechanism is available to link from Cordis to OpenAire, the RCN number. The OpenAire catalogue can be queried using an RCN filter to retrieve only resources relevant to a project. This work is still in preparation.
Not all DOI's registered in Cordis are available in OpenAire. OpenAire only lists resources with an open access license. Other DOI's can be fetched from the DOI registry directly or via Crossref.org. This work is still in preparation. Detailed technical information can be found in the technical description.
OpenAire and other sources
The software used to query OpenAire by DOI or by RCN is not limited to be used by DOIs or RCNs that come from Cordis. Any list of DOIs or list of RCNs can be handled by the software.
Risks & Limitations
Repository Storage
Introduction
Overview and Scope
The SoilWise repository aims at merging and seamlessly providing different types of content. To host this content and to be able to efficiently drive internal processes and to offer performant end user functionality, different storage options are implemented.
- A relational database management system for the storage of the core metadata of both data and knowledge assets.
- A Triple Store to store the metadata of data and knowledge assets as a graph, linked to soil health and related knowledge as a linked data graph.
- Git for storage of user-enhanced metadata.
Intended Audience
Storage is a backend component, therefore we only expect a maintenance role:
- SWC Administrator monitoring the health status, logs, signaling maintenance issues etc .
- SWC Maintainer performing corrective / adaptive maintenance tasks that require database access and updates.
PostgreSQL RDBMS: storage of raw and augmented metadata
Info
Current version: Postgres release 12.2;
Technology: Postgres
Access point: SQL
A "conventional" RDBMS is used to store the (augmented) metadata of data and knowledge assets. There are several reasons for choosing an RDBMS as the main source for metadata storage and metadata querying:
- An RDBMS provides good options to efficiently structure and index its contents, thus allowing performant access for both internal processes and end user interface querying.
- An RDBMS easily allows implementing constraints and checks to keep data and relations consistent and valid.
- Various extensions, e.g. search engines, are available to make querying, aggregations even more performant and fitted for end users.
Key Features
The Postgres database serves as a the destination and/or source for many of the backend processes of the SoilWise Catalogie. Its key features are:
- Raw metadata storage — The harvester process uses it to store the raw results of the metadata harvesting of the different resources that are currently connected.
- Storage of Augmented metadata — Various metadata augmentation jobs use it as input and write their input to this data store.
- Source for Search Index processing — This database is also the source for denormalisation, processing and indexing metadata through the Solr framework.
- Source for UI querying — While Solr is the main resource for end user querying through the catalogue UI, the catalogue also queries the Postgress database.
Virtuoso Triple Store: storage of SWR knowledge graph
Info
Current version: Virtuoso release 07.20.3239
Technology: Virtuoso
Access point: Triple Store (SWR SPARQL endpoint) https://repository.soilwise-he.eu/sparql
A Triple Store is implemented as part of the SWR infrastructure to allow a more flexible linkage between the knowledge captured as metadata and various sources of internal and external knowledge sources, particularly taxonomies, vocabularies and ontologies that are implemented as RDF graphs. Results of the harvesting and metadata augmentation that are stored in the RDBMS are converted to RDF and stored in the Triple Store.
Key Features
A Triple Store, implemented in Virtuoso, is integrated for parallel storage of metadata because it offers several capabilites:
- Semantic linkage — It allows the linking of different knowledge models, e.g. to connect the SWR metadata model with existing and new knowledge structures on soil health and related domains.
- Cross-domain reasoning — It allows reasoning over the relations in the stored graph, and thus allows connecting and smartly combining knowledge from those domains.
- Semantic querying — The SPARQL interface offered on top of the Triple Store allows users and processes to use such reasoning and exploit previously unconnected sets of knowledge.
Apache Lucene: Open-source search engine software library
Info
Current version: Apache Lucene release x.x.x
Technology: Apache Lucene
Access point: Via the Apache Solr API https://
The SoilWise Catalogie uses a dedicated index (Apache Lucene) to efficiently index and store the harvested and augmented metadata, as well as the knowledge extracted from documents referred to through the metadata records (currently only supporting PDF format). Access to the index (both indexing and querying) is provided through the Apache Solr search framework.
Key Features
Apache Lucene offers a range of options that support increasing the search performance and the quality of search results. It also allows to implement strategies for result ranking, faceted search etc. that can increase end user experience :
- Search performance — Apache Lucene is a broadly adopted and well maintaned search index that can dramatically speed up and improve the precision of search results
- Integration — Integration with Apache Solr provides tools and programmatic access to configure, manage, optimize and query the indexed content.
- Lexical Search — Combined with the Apache Solr framework, Lucene offers support for lexical search (based on matching the literals of words and their variants), faceted search and ranking.
- Semantic Search — Combined with the Apache Solr framework, Lucene offers support for generating and storing embeddings that support semantic search (based on the meaning of data) and associated AI functions.
Git: storage of code and configuration
Info
Technology: Gitlab and GitHub
Access point: https://github.com/soilwise-he
Git is a multi purpose environment for storing and managing software and documentation, versioning and configuration that also offers various functions the support the management and monitoring of the software development process.
Key Features
Git is an acknowledged platform to store, version, configure and docuemnt software, with additional features for software and software development management. The key features used in SoilWise are:
- Code storage, version and configuration management — Git is used to deposit and manage versions of Soilwise code, documentation and configurations.
- Issue and release management — SoilWIse uses the issue and release management to document, monitor and track the development of software conponents and their integration.
- Process automation — Git defines and runs automated pipelines for deployment, augmentation, validation and harvesting external sources.
Integrations & Interfaces
Key Architectural Decisions
| Decision | Rationale |
|---|---|
| RDF/Triple Store for semantics | Allows definition of advanced semantic structures and cross-domain interlnkage. Allows semantic reasoning, both internal and by external clients |
| To be further extended | ... |
Risks & Limitations
| Risk / Limitation | Description | Mitigation |
|---|---|---|
| Inconsistency between RDBMS and Triple Store | Parallel sources and query results might deviate if processes are not aligned. | Monitoring procedures and corrective actions to be documented for maintenance |
| Integration issues for Triple store | Lack of infrastructure and/or technical knowledge might hinder integration. | Continuous alignment with JRC technical team |
| Integration issues for process automation | Currently implemented process automation through Git might not fit JRC | Continuous alignment with JRC technical team |
| To be further extended | ... |
Metadata Catalogue
Info
Current version:
Technology: Apache Solr, React, pycsw
Project: Catalogue UI; Solr; pycsw
Access point: https://client.soilwise-he.containers.wur.nl/catalogue/search
Introduction
Overview and Scope
The Metadata Catalogue is a central piece of the architecture, giving access to individual metadata records. In the catalogue domain, various effective metadata catalogues are developed around the standards issued by the OGC, the Catalogue Service for the Web (CSW) and the OGC API Records, Open Archives Initiative (OAI-PMH), W3C (DCAT), FAIR science (Datacite) and Search Engine community (schema.org). For our first project iteration we've selected the pycsw software, which supports most of these standards. In the second iteration pycsw continues to provide standardized APIs, however to improve search performance and user experience, it was supplemented by Apache Solr and React frontend.
Intended Audience
The SoilWise Metadata Catalogue targets the following user groups:
- Soil scientists and researchers working with European soil health data and seeking catalogued knowledge, publications, and datasets.
- Living Labs' data scientists working with European soil health data and seeking catalogued knowledge, publications, and datasets.
- Mission Soil Project Data Managers searching for datasets published by other Mission Soil Projects, or veryfiing if their published data are recognized by SoilWise (EUSO).
- Policy Makers working with European soil health data and seeking catalogued knowledge, publications, and datasets.
Key Features
User interface
The SoilWise Metadata Catalogue adopts a React frontend, focusing on:
- Paginated search results - Search results are displayed per page in ranked order, in the form of overview table comprising preview of resource type, title, abstract, date and preview.
- Fulltext search - + autocomplete
- Resource type filter - enabling to filter out certain types of resources, e.g. journal articles, datasets, reports, software.
- Term filter - enabling to filter out resources containing certain keywords, resources published by specific projects, etc.
- Date filter - enabling to filter out resources based on their creation, or revision date
- Spatial filter - enabling to filter out resources based on their spatial extent using countries or regions, drawn bounding box, vicinity of user's location, or by searching for geographical names.
- Record detail view - After clicking result's table item, a record's detail is displayed at unique URL address to facilitate sharing. Record's detail currently comprises: record's type tag,full title, full abstract, keywords' tags, preview of record's geographical extent, record's preview image, if available, information about relevant HE funding project, list of source repositories,- indication of link availability, see Link liveliness assessment, last update date, all other record's items...
- Resource preview - 3 types of preview are currently supported: (1) Display resource geographical extent, which is available in the record's detail, as well in the search results list. (2) Display of a graphic preview (thumbnail) in case it is advertised in metadata. (3) Map preview of OGC:WMS services advertised in metadata enables standard simple user interaction (zoom, changing layers).
- Display results of metadata augmentation - Results of metadata augmentation are stored on a dedicated database table. The results are merged into the harvested content during publication to the catalogue. At the moment it is not possible to differentiate between original and augmented content. For next iterations we aim to make this distinction more clear.
- Display links of related information - Download of data "as is" is currently supported through the links section from the harvested repository. Note, "interoperable data download" has been only a proof-of-concept in the first iteration phase, i.e. is not integrated into the SoilWise Catalogue. Download of knowledge source "as is" is currently supported through the links section from the harvested repository.
Index and search strategies
The SoilWise Metadata Catalogue implements back-end index and search functions based on Apache Solr, focusing on:
- Denormalising metadata - Solr is set up as a document indexing infrastructure, working on rather "flat" textual formats instead of normalised database models. The first step is therefore a conversion to a denormalised structure, currently implemented as a (single) database view.
- Composing Solr documents - From the denormalised view, the individual metadata records are processed into Solr.documents.
- Transforming/Indexing - Solr uses transformers to process and index Solr.documents. This is a combination of sequential sub processes (e.g. tokenizers) and configurations that determine how the documents are indexed and how they can be searched, ranked, feceted etc.
- Search API - The Solr search API Allows query access to the Solr index, so the UI (and other clients) can search the metadata through the index.
Supported standards
In order to interact with the many relevant data communities, SoilWise aims to support a range of catalogue standards through pycsw backend, for more info see Integrations & Interfaces.
Architecture
Technological Stack
Backend
| Technology | Description |
|---|---|
| pycsw v3.0 | Pycsw, written in python, allows for the publishing and discovery of geospatial metadata via numerous APIs (CSW 2/CSW 3, OAI-PMH, providing a standards-based metadata and catalogue component of spatial data infrastructures. pycsw is Open Source, released under an MIT license, and runs on all major platforms (Windows, Linux, Mac OS X). |
| Apache Lucene v11.x | Apache Lucene is a open source high-performance Java-based search engine library. |
| Apache Solr v9.7.0 | Open source full text, vector and geo-spatial search framework on top of the Apache Lucene Index. |
| Java vx.x | |
| OpenStreetMaps API |
Frontend
| Technology | Description |
|---|---|
| React | Javascript framework that implements the search interface and access to Solr API |
| pycsw v3.0 | (depricated) Pycsw also offers its own User interface, which was used as a default in previous SoilWise prototype. |
Infrastructure
| Component | Technology |
|---|---|
| Container | Docker (multi-stage build, Eclipse Temurin JDK 21) |
| CI/CD | GitLab CI with semantic release (conventional commits) |
| Orchestration | Kubernetes (liveness/readiness probes) |
Main Component Diagram
Main Sequence Diagram
Integrations & Interfaces
| Service | Auth | Endpoint | Purpose |
|---|---|---|---|
| Catalogue Service for the Web (CSW) | https://repository.soilwise-he.eu/cat/csw | Catalogue service for the web (CSW) is a standardised pattern to interact with (spatial) catalogues, maintained by OGC. | |
| OGC API - Records | https://repository.soilwise-he.eu/cat/openapi | OGC is currently in the process of adopting a revised edition of its catalogue standards. The new standard is called OGC API - Records. OGC API - Records is closely related to Spatio Temporal Asset Catalogue (STAC), a community standard in the Earth Observation community. | |
| Protocol for metadata harvesting (OAI-PMH) | https://repository.soilwise-he.eu/cat/oaipmh | The open archives initiative has defined a common protocol for metadata harvesting (oai-pmh), which is adopted by many catalogue solutions, such as Zenodo, OpenAire, CKAN. The oai-pmh endpoint of Soilwise can be harvested by these repositories. | |
| Spatio Temporal Asset Catalog (STAC) | https://repository.soilwise-he.eu/cat/stac/openapi | ||
| OpenSearch | https://repository.soilwise-he.eu/cat/opensearch |
Key Architectural Decisions
Risks & Limitations
| Risk / Limitation | Description | Mitigation |
|---|---|---|
| Transferability | The differences in technology stack between the implementing consortium and the final owner (JRC) might lead to transferability and integration issues | Use of broadly adopted open source products. Alignment with JRC technical team |
| Metadata quality | The performance of the search functionality is highly dependent on the completeness and quality of the harvested metadata which is out of scope for SoilWise. | Metadata augmnentation will allow to partly mitigate |
| Transparency and explainability | The dependency on metadata completeness and quality in combination with the large amount of interdependent options for (fuzzy) search strategies and the different combinations of UI search features will make it hard to understand the logic behind search results. | Documentation of metadata augmentation, search strategies etc. |
| Usability | The diversity of user groups and their requirements and expectations make it difficult to find balance between functionality/complexity/user-friendliness. | Iterative appraoch and validation/testing with user groups to align. |
Soil Companion (chatbot)
Info
Current version: 1.2.x
Technology: Retrieval Augmented Generative Artificial Intelligence
Project: Soil Companion
Access point: https://soil-companion.containers.wur.nl/app/index.html
Introduction
Overview and Scope
The Soil Companion is an AI chatbot developed in the SoilWise project. It provides an intelligent conversational interface through which users can explore European soil metadata, query global and country-specific soil data services, and receive answers grounded in the SoilWise knowledge repository.
The chatbot uses an agentic tool-calling approach: a large language model (LLM) autonomously decides which external data sources to consult for each question, executes the relevant tool calls, and synthesizes the results into a coherent response. Answers are enriched with auto-generated links to SoilWise vocabulary terms and Wikipedia articles. A sidebar Insight panel displays related SKOS vocabulary concepts and clickable chips that allow users to explore connected topics.
Intended Audience
The Soil Companion targets the following user groups:
- Soil scientists and researchers working with European soil health data and seeking catalogued knowledge, publications, and datasets from the SoilWise repository.
- Agricultural experts and extension officers looking for soil property data, field-level KPIs, and crop information to inform land management decisions.
- Students and educators exploring soil science concepts through a conversational interface that provides definitions, vocabulary hierarchies, and links to authoritative sources.
- Farmers and land managers (in selected regions) who want accessible field-level agricultural data such as crop history, soil physical properties, and greenness indices.
Key Features
The chatbot combines agentic LLM tool calling with retrieval-augmented generation and post-response enrichment to deliver grounded, linked answers. The key features are:
- Agentic tool calling — The LLM autonomously decides which of the available tool integrations to invoke (catalog search, SoilGrids, AgroDataCube, Wikipedia, vocabulary SPARQL), executing up to 10 sequential tool-call iterations per query.
- RAG from local core knowledge — Documents (PDF, text, Markdown) are split into chunks, embedded with a local model (AllMiniLmL6V2), and stored in memory. Relevant chunks are retrieved by cosine similarity and injected into the prompt.
- Response enrichment — After the LLM generates a response, auto-linkers scan for vocabulary terms and Wikipedia article titles, inserting navigable links into the rendered output.
- Insight panel — The frontend extracts SoilWise and Wikipedia links from responses and displays broader/narrower/related vocabulary concepts with definitions in a sidebar panel.
- Token streaming — Responses are streamed token-by-token over WebSocket, giving users immediate visual feedback.
- Feedback loop — Thumbs up/down ratings are logged to daily JSONL files; evaluation tools compute quality metrics (like rate, NSAT, Wilson lower bound).
Architecture
Technological Stack
Backend (JVM)
| Component | Technology |
|---|---|
| Language | Scala 3.8.x on JDK 17+ (tested 17–25) |
| Build | SBT 1.11.x / 1.12.x (cross-build JS/JVM) |
| LLM Framework | LangChain4j 1.10.x (OpenAI integration, agentic tool calling, embeddings, RAG) |
| LLM Provider | OpenAI (gpt-4o-mini for chat, gpt-4o for reasoning, text-embedding-3-small for embeddings) |
| Local Embeddings | AllMiniLmL6V2 (offline, ~33 MB model for RAG document retrieval) |
| Vector Store | In-memory embedding store (with experimental Chroma support) |
| Logging | SLF4J 3.0.x + Logback 1.5.x (daily rotation, 30-day retention) |
| Document Parsing | Apache Tika (PDF, text, Markdown) |
Frontend (Browser)
| Component | Technology |
|---|---|
| Language | Scala.js (compiled to JavaScript) |
| Maps | Leaflet 1.9.x |
| Communication | WebSocket (real-time streaming) |
Infrastructure
| Component | Technology |
|---|---|
| Container | Docker (multi-stage build, Eclipse Temurin JDK 21) |
| CI/CD | GitLab CI with semantic release (conventional commits) |
| Orchestration | Kubernetes (liveness/readiness probes) |
Main Components Diagram
High-level component overview:
graph TD
subgraph Browser ["Browser (Scala.js)"]
App["SoilCompanionApp
- WebSocket client (real-time chat streaming)
- Authentication & session management
- Location picker (Leaflet map)
- Insight panel (vocabulary concepts, Wikipedia links)
- File upload, feedback, theme toggle"]
end
Browser -- "WebSocket + HTTP" --> Server
subgraph Server ["SoilCompanionServer (Cask, JVM)"]
direction TB
Routes["Routes: /healthz, /readyz, /login, /logout, /session,
/subscribe/:id (WS), /query, /clear, /upload,
/feedback, /location, /vocab, /app/*"]
subgraph Internal_Modules [" "]
direction LR
Config["Config
(PureConfig)"]
Logger["Feedback
Logger"]
SessMgmt["Session Management
(ConcurrentHashMaps)"]
end
subgraph Assistant ["Assistant (per session)"]
AIServices["LangChain4j AiServices
- StreamingChatModel (OpenAI)
- ChatMemory (50 messages)
- RAG ContentRetriever (embeddings + local docs)
- Tool methods (5 integrations)"]
end
subgraph PostLinking ["(post-response linking)"]
direction LR
VL[VocabLinker]
WL[WikipediaLinker]
end
end
Server -- "HTTP calls" --> External
subgraph External ["External Services"]
direction TB
E1["OpenAI API
(LLM, embed)"]
E2["Solr
(catalog)"]
E3["ISRIC SoilGrids v2.0
(global soil properties)"]
E4["SoilWise
SPARQL"]
E5["Wikipedia
(6 langs)"]
E6["WUR AgroDataCube v2
(NL field data)"]
end
%% Styling
style Browser fill:#f9f9f9,stroke:#333,stroke-width:2px
style Server fill:#fff,stroke:#333,stroke-width:2px
style External fill:#f9f9f9,stroke:#333,stroke-width:2px
style Assistant fill:#fff,stroke:#333,stroke-dasharray: 5 5
style Internal_Modules fill:none,stroke:none
style PostLinking fill:none,stroke:none
Main Sequence Diagram
User query to response flow:
sequenceDiagram
autonumber
participant C as Client (Browser)
participant S as Server (JVM)
participant E as External APIs
Note over C, S: Session Initialization
C->>S: GET /session
S-->>C: { sessionId: UUID }
C->>S: WS /subscribe/:sessionId
Note right of S: store connection
S-->>C: connection established
C->>S: POST /login
Note right of S: validate credentials
S-->>C: { ok: true }
Note over C, S: Chat Interaction
C->>S: POST /query
{ sessionId, content }
S-->>C: QueryEvent("received")
Note right of S: generate questionId
S-->>C: QueryEvent("thinking")
S-->>C: QueryEvent("retrieving_context")
S->>E: RAG: embed query
E-->>S: top-5 document chunks
S->>E: LLM: evaluate tools (OpenAI)
E-->>S: tool call decision
S->>E: Tool: e.g. Solr search (Solr)
E-->>S: search results
S->>E: LLM: synthesize answer (OpenAI)
E-->>S: token stream begins
loop Token Streaming
S-->>C: QueryPartialResponse(token)
end
Note right of S: stream complete
Apply VocabLinker
Apply WikipediaLinker
S-->>C: QueryEvent("links_added", linkedResponse)
S-->>C: QueryEvent("done")
Note left of C: render markdown,
show feedback buttons
C->>S: POST /feedback
{ questionId, vote }
Note right of S: log to JSONL
Database Design
The Soil Companion does not use a traditional database. All runtime state is held in-memory; only feedback and application logs are persisted to disk.
In-memory state (per server process):
| Store | Type | Purpose |
|---|---|---|
wsConnections |
ConcurrentHashMap[String, WsChannelActor] |
Active WebSocket connections |
assistants |
ConcurrentHashMap[String, Assistant] |
LLM chat state per session |
uploadedTexts |
ConcurrentHashMap[String, String] |
Temporary uploaded file content |
uploadedFilenames |
ConcurrentHashMap[String, String] |
Original filenames of uploads |
locationContexts |
ConcurrentHashMap[String, String] |
Location JSON per session |
authenticatedSessions |
ConcurrentHashMap[String, Boolean] |
Authentication status |
lastActivity |
ConcurrentHashMap[String, Long] |
Session inactivity tracking |
In-memory vector store:
| Store | Type | Purpose |
|---|---|---|
embeddingStore |
InMemoryEmbeddingStore[TextSegment] |
Embedded knowledge document chunks |
Documents from the data/knowledge/ directory are loaded, split into 500-character chunks (100-character overlap), embedded using AllMiniLmL6V2, and stored at startup.
Persistent file storage:
| Data | Location | Format |
|---|---|---|
| Feedback | data/feedback-logs/feedback-YYYY-MM-DD.jsonl |
Daily JSONL, auto-rotated, gzip compressed |
| Application logs | data/logs/soil-companion.log |
Logback rolling file (30-day retention, gzip) |
| Knowledge documents | data/knowledge/ |
PDF, text, Markdown (read-only at startup) |
| Vocabulary | data/vocab/soilvoc_concepts_*.csv |
CSV (loaded at startup for auto-linking) |
Integrations & Interfaces
| Service | Auth | Endpoint | Purpose |
|---|---|---|---|
| OpenAI API | Bearer token (OPENAI_API_KEY) |
via LangChain4j | Chat completion (gpt-4o-mini), reasoning (gpt-4o), embeddings (text-embedding-3-small) |
| Solr (SoilWise Catalog) | Basic Auth (SOLR_USERNAME / SOLR_PASSWORD) |
SOLR_BASE_URL |
Search datasets and publications; full-text content retrieval |
| ISRIC SoilGrids v2.0 | None (public) | SOILGRIDS_BASE_URL |
Soil property estimates at lat/lon (~250 m resolution) |
| SoilWise SPARQL | None | VOCAB_SPARQL_ENDPOINT |
SKOS concept hierarchies (broader, narrower, related terms) |
| Wikipedia | None (public) | WIKIPEDIA_BASE_URL (per language) |
Article search and content retrieval (6 languages) |
| WUR AgroDataCube v2 | Token header (AGRODATACUBE_ACCESS_TOKEN) |
AGRODATACUBE_BASE_URL |
NL field parcels, crop history, soil/crop KPIs |
All external service credentials and endpoints are configured through HOCON (application.conf) with environment variable overrides.
HTTP endpoints exposed by the server:
| Method | Path | Purpose |
|---|---|---|
GET |
/healthz |
Liveness probe (version, uptime) |
GET |
/readyz |
Readiness probe (config + API key checks) |
POST |
/login |
Demo authentication |
POST |
/logout |
Session teardown |
GET |
/session |
New session ID |
WS |
/subscribe/:sessionId |
WebSocket for streaming chat |
POST |
/query |
Submit a question |
POST |
/clear |
Clear chat history |
POST |
/upload |
Upload text/Markdown context |
POST |
/feedback |
Submit thumbs up/down |
POST |
/location |
Set geographic context |
POST |
/vocab |
Batch vocabulary concept lookup |
GET |
/app/* |
Static frontend assets |
WebSocket event types:
| Event | Direction | Purpose |
|---|---|---|
received |
Server → Client | Query acknowledged, questionId assigned |
thinking |
Server → Client | LLM is analysing the question |
retrieving_context |
Server → Client | RAG retrieval in progress |
generating |
Server → Client | LLM is generating the answer |
links_added |
Server → Client | Auto-linked response replacing the streamed version |
done |
Server → Client | Response complete |
error |
Server → Client | An error occurred |
heartbeat |
Server → Client | Keep-alive (every 15 seconds) |
session_expired |
Server → Client | Session timed out due to inactivity |
prompt_truncated |
Server → Client | Input was truncated to stay within limits |
QueryPartialResponse |
Server → Client | Single streamed token |
Key Architectural Decisions
| Decision | Rationale |
|---|---|
| Scala 3 + Scala.js cross-build | Enables shared domain models (QueryRequest, QueryEvent, QueryPartialResponse) between backend and frontend, eliminating serialization mismatches and reducing code duplication. |
| Cask HTTP micro-framework | Lightweight, Scala-native server with built-in WebSocket support. Suitable for a single-service chatbot without the overhead of a full application framework. |
| LangChain4j for LLM integration | Provides a mature JVM-native abstraction for tool calling, RAG, streaming, and chat memory — avoiding the need to call OpenAI APIs directly. The @Tool annotation enables declarative tool registration. |
| In-memory embedding store | Simplifies deployment (no external vector database required). Sufficient for the current knowledge base (~5 documents). Trade-off: state is lost on restart and capacity is limited by server memory. |
| AllMiniLmL6V2 for local embeddings | Runs offline without API calls, keeping RAG retrieval fast and cost-free. The ~33 MB model is small enough to bundle in the Docker image. |
| Per-session Assistant instances | Each session gets its own Assistant with isolated chat memory, tool state (e.g. AgroDataCube field context), and uploaded file context. Prevents cross-session contamination. |
| Post-response auto-linking | VocabLinker and WikipediaLinker run after the LLM completes, replacing the streamed response with an enriched version. This avoids asking the LLM to generate links (which is unreliable) while still providing navigable references. |
| WebSocket token streaming | Provides immediate visual feedback during LLM generation, reducing perceived latency. A 15-second heartbeat prevents proxy/ingress idle timeouts. |
| Environment variable configuration | All credentials and endpoints are overridable via environment variables, following 12-factor app principles for containerized deployment. |
| Demo authentication | A simple single-user mode enables local development and demonstrations without requiring an external identity provider. Production deployment would integrate with an external auth layer. |
Risks & Limitations
| Risk / Limitation | Description | Mitigation |
|---|---|---|
| SoilGrids accuracy | Returned values are modelled estimates at ~250 m grid resolution, not field measurements. | Tool responses include explicit disclaimers advising users to verify with local data. |
| Single-user demo auth | The demo authentication mode uses a single configurable account with no roles or authorization. Not suitable for production multi-user scenarios. | Designed for development/testing; production deployment requires integration with an external authentication and authorization layer. |
| In-memory state loss | All session state, chat memory, uploaded context, and the embedding store are lost on server restart. | Acceptable for a demo/chatbot use case. Persistent vector store (Chroma) support exists experimentally for future use. |
| No horizontal scaling | All sessions are held in a single JVM process with no shared session store. | Sufficient for current usage levels. Horizontal scaling would require an external session store and load balancer. |
| External API availability | The chatbot depends on multiple external APIs (Solr, SoilGrids, AgroDataCube, OpenAgroKPI, Wikipedia). Downtime or rate limits on any service degrades functionality. | Tool methods handle errors gracefully, returning informative messages. The LLM can fall back to other tools if one fails. LangChain4j provides configurable retry logic (max 3 retries). |
| Geographic coverage | AgroDataCube and OpenAgroKPI are Netherlands-only. SoilGrids is global but at coarse resolution. | Tool descriptions inform the LLM of geographic scope so it can communicate limitations to users. |
| Knowledge base is static | Local documents are loaded and embedded only at startup. No hot-reload mechanism exists. | A server restart picks up new documents. This is acceptable for infrequently changing knowledge resources. |
| LLM hallucination | Despite RAG grounding and tool results, the LLM may still generate inaccurate statements. | System prompts instruct the model to include disclaimers and prefer tool-grounded answers. User feedback collection enables ongoing quality monitoring. |
| Prompt injection via uploads | Uploaded files and location contexts could contain adversarial content. | Input sanitization is applied; prompt size is capped at 120,000 characters; file uploads are limited to 200 KB. |
| CORS policy | The file upload endpoint uses a permissive Access-Control-Allow-Origin: * header. |
Acceptable for demo deployment; should be tightened for production. |
Data publication support
Introduction
Overview and Scope
At the moment, SoilWise supports data publishers with the following tools:
- DOI Resolution Widget
- Tabular Soil Data Annotation to help users create semantic metadata for tabular datasets.
- INSPIRE Geopackage Transformation
- Soil Vocabulary Viewer, part of the Knowledge Graph component, visualizes and links different soil-domain vocabularies and terms.
Intended Audience
DOI Resolution Widget
Info
Current version:
Technology:
Project:
Access Point:
Overview and Scope
Key Features
Architecture
Technological Stack
Main Sequence Diagram
Integrations & Interfaces
Key Architectural Decisions
Risks & Limitations
Tabular Soil Data Annotation
Info
Current version:
Technology: Streamlit, Python, OpenAI API
Project: Tabular Data Annotator
Access Point: https://dataannotator-swr.streamlit.app/
Overview and Scope
DataAnnotator is a Streamlit-based web application designed to help users create semantic metadata for tabular datasets. It combines optional Large Language Model (LLM) assistance with semantic embeddings to annotate data columns with machine-readable descriptions, element definitions, units, methods, and vocabulary mappings.
The tool addresses the metadata annotation workflow by:
- Enabling manual annotation: Users directly enter descriptions for data columns
- Automating description generation (optional): If users have context documentation, LLMs can help extract and structure descriptions automatically
- Linking to vocabularies: Semantic embeddings match descriptions to controlled vocabularies for standardization
The LLM layer is optional—users can skip automated generation and manually provide descriptions, which the system will then semantically match to existing vocabulary terms.
Key Features
| Feature | Implementation | Purpose |
|---|---|---|
| Auto Type Detection | Statistical sampling | Identify data patterns |
| Manual Description Entry | Streamlit text inputs | Direct user annotation |
| Optional LLM Assistance | OpenAI/Apertus integration | Auto-extract descriptions from docs |
| Semantic Vocabulary Matching | FAISS vector search | Link descriptions to standard vocabularies |
| Context Awareness | PDF/DOCX import + prompting | Extract domain-specific info when available |
| Multi-format Export | flat csv/TableSchema/CSVW | Integration with downstream tools |
Architecture
Technological Stack
| Component | Technology |
|---|---|
| Frontend | Streamlit 1.51+ |
| Backend Logic | Python 3.12+ |
| LLM Integration | OpenAI API, Apertus HTTP |
| Embeddings | Sentence Transformers 5.1+ |
| Vector Search | FAISS (CPU) 1.12+ |
| File Parsing | PyPDF2, python-docx, openpyxl |
| ML Libraries | scikit-learn, NumPy |
Dependencies & Models
-
Python Packages
streamlit>=1.51.0- Web UI frameworkopenai>=2.7.2- LLM API clientsentence-transformers>=5.1.2- Semantic embeddingfaiss-cpu>=1.12.0- Vector similarity searchpandas>=2.0- Data manipulationopenpyxl>=3.1.5- Excel handlingpython-docx>=1.2.0- Word document parsingpypdf2>=3.0.1- PDF text extraction
-
Pre-trained Models/Data
- Embedding Model:
all-MiniLM-L6-v2(384 dimensions, 22M parameters) - FAISS Index: Pre-computed vocabulary embeddings (stored in
data/directory)
- Embedding Model:
Main Components
1. Data Input Module
- Supported Formats - Tabular Data: CSV, Excel (XLSX)
- Supported Formats - Context Documents: Free-form text input, PDF documents, DOCX files
- Processing Functions:
read_csv_with_sniffer(): Auto-detects CSV delimitersimport_metadata_from_file(): Reads existing metadata if providedread_context_file(): Extracts context from PDFs/DOCX for LLM-assisted annotation
2. Data Analysis & Type Detection
- Function:
detect_column_type_from_series() - Detects:
- String: Text data
- Numeric: Integers and floats
- Date: Temporal values
- Approach: Statistical sampling of column values (up to 200 non-null entries)
3. Metadata Framework
- Function:
build_metadata_df_from_df() - Template Fields:
name: Column identifierdatatype: Type classification (string/numeric/date)element: Semantic element definitionunit: Measurement unitmethod: Collection/calculation methoddescription: Human-readable descriptionelement_uri: Link to external vocabulary
4. LLM Integration Layer (Optional)
Purpose: Automate the extraction and structuring of descriptions from existing documentation when users have context materials.
When to Use: - User has documentation (PDFs, Word docs, etc.) describing variables - Manual annotation is time-consuming for large datasets - Descriptions need to be extracted from unstructured text
Supported Providers:
-
OpenAI (Recommended)
- Uses GPT models for high-quality response generation
get_response_OpenAI(): Direct API calls- Best for complex, domain-specific text extraction
-
Apertus (Alternative)
- Self-hosted LLM option
get_response_Apertus(): HTTP-based endpoint- Swiss-based open-source model
Functionality:
- Function:
generate_descriptions_with_LLM() - Inputs:
- Variable names to describe
- Context information from documents or text input
- Output Format: Structured JSON with descriptions for each variable
5. Semantic Embedding & Vocabulary Matching
- Model: Sentence Transformers (default:
all-MiniLM-L6-v2) - Functions:
load_sentence_model(): Load embedding modelload_vocab_indexes(): Load pre-computed FAISS indexesembed_vocab(): Generate embeddings with optional definition weighting
- Purpose: Match generated or manually-entered descriptions to controlled vocabularies
Pre-computed Vocabulary Sources
The FAISS vectorstore was pre-computed by embedding terms from four major public vocabularies:
| Vocabulary | Domain | Source |
|---|---|---|
| Agrovoc | Agricultural and food sciences | FAO - Food and Agriculture Organization |
| GEMET | Environmental terminology | European Environment Agency (EEA) |
| GLOSIS | Soil science and properties | FAO Global Soil Information System |
| ISO 11074:2005 | Soil quality terminology | International Organization for Standardization |
This multi-vocabulary approach enables annotation of diverse datasets including agricultural, environmental, and soil-related data.
FAISS Index Structure:
Index File: vocabCombined-{modelname}.index
Metadata File: vocabCombined-{modelname}-meta.npz
Metadata Dictionary Format:
{
index_id: {
"uri": "vocabulary_uri",
"label": "preferred_label",
"definition": "term_definition",
"QC_label": "prefLabel|altLabel"
},
...
}
6. Export & Download Module
- Function:
download_bytes() - Supported Formats:
- Excel (XLSX) - for human review
- JSON - for machine processing
- CSV - for spreadsheet tools
- Implementation: Streamlit session-based download management
Main Sequence Diagram
graph TB
A["User Interface
Streamlit App"] -->|Upload Data| B["Data Input Handler
CSV/Excel/PDF"]
B -->|Parse Data| C["Data Analysis
Column Type Detection"]
C -->|Analyze Structure| D["Metadata Framework
Build Template"]
D --> E{"Description Source?"}
E -->|Manual Entry| F["User Provides
Descriptions"]
E -->|Optional: Auto-generate| I[/"LLM Provider
(Optional Tool)"\]
I -->|OpenAI API| J["OpenAI Client
GPT Models"]
I -->|Apertus API| K["Apertus Client
Local LLM"]
J -->|JSON Descriptions| L["Response Parser
JSON Extraction"]
K -->|JSON Descriptions| L
L --> M["LLM-generated
Descriptions"]
F --> N["Semantic Embedding
Sentence Transformers"]
M --> N
N -->|Vector Query| O["proposal for generalized ObservedProperty"]
V1["Agrovoc
Agricultural"] -.->|Pre-embedded| G["FAISS vector store"]
V2["GEMET
Environmental"] -.->|Pre-embedded| G
V3["GLOSIS
Soil Science"] -.->|Pre-embedded| G
V4["ISO 11074:2005
Soil Quality"] -.->|Pre-embedded| G
G --> O
O -->Q["Export Handler
flat csv/TableSchema/CSVW"]
Q -->|Downloaded metadata| A
Key Architectural Decisions
Optimization Strategies:
- Model Caching: Streamlit
@st.cache_resourcefor persistent model loading - API Caching: JSON-based result memoization to avoid redundant API calls
- FAISS Optimization: Pre-computed indexes for O(log n) vector search
- Batch Processing: Process multiple columns in single LLM call
INSPIRE Geopackage Transformation
Info
Current version:
Technology:
Project:
Access Point:
Overview and Scope
Key Features
User Manual
Architecture
Technological Stack
Main Sequence Diagram
Database Design
Integrations & Interfaces
Key Architectural Decisions
Risks & Limitations
Other recommended tools acknowledged by SoilWise community
The following components are not a product of SoilWise project, and not an integral part of the SoilWise Catalogue, but are recommended by the SoilWise community.
Hale Studio
A proven ETL tool optimised for working with complex structured data, such as XML, relational databases, or a wide range of tabular formats. It supports all required procedures for semantic and structural transformation. It can also handle reprojection. While Hale Studio exists as a multi-platform interactive application, its capabilities can be provided through a web service with an OpenAPI.
User Manual
A comprehensive tutorial video on soil data harmonisation with hale studio can be found here.
Setting up a transformation process in hale»connect
Complete the following steps to set up soil data transformation, validation and publication processes:
- Log into hale»connect.
- Create a new transformation project (or upload it).
- Specify source and target schemas.
- Create a theme (this is a process that describes what should happen with the data).
- Add a new transformation configuration. Note: Metadata generation can be configured in this step.
- A validation process can be set up to check against conformance classes.
Executing a transformation process
- Create a new dataset and select the theme of the current source data, and provide the source data file.
- Execute the transformation process. ETF validation processes are also performed. If successful, a target dataset and the validation reports will be created.
- View and download services will be created if required.
To create metadata (data set and service metadata), activate the corresponding button(s) when setting up the theme for the transformation process.
Metadata Validation
Introduction
Metadata should help users assess the usability of a data set for their own purposes and help users to understand their quality. Assessing the quality of metadata may guide the stakeholders in future governance of the system.
Overview and Scope
In terms of metadata, SoilWise Repository aims for the approach to balance harvesting between quantity and quality. See for more information in the Harvester Component. Catalogues which capture metadata authored by various data communities typically have a wide range of metadata completion and accuracy. Therefore, the SoilWise Repository employs metadata validation mechanisms to provide additional information about metadata completeness, conformance and integrity. Information resulting from the validation process are stored together with each metadata record in a relation database and updated after registering a new metadata version. Within the first iteration, they are not displayed in the SoilWise Catalogue.
On this topic 2 components are available which monitor aspects of metadata
- Metadata completeness calculates a score based on selected populated metadata properties
- Metadata INSPIRE compliance
- Link Liveliness Assessment part of Metadata Augmentation components, validates the availability of resources described by the record
Intended Audience
The SoilWise Metadata Validation targets the following user groups:
- JRC data analysts monitoring the metadata quality of published soil-related datasets.
Metadata completeness
Info
Current version: 0.2.0
Technology: Python
Project: Metadata validator
Access point: Postgres database
The software calculates a level of completeness of a record, indicated in % of 100 for endorsed properties, considering that some properties are conditional based on selected values in other properties.
Completeness is evaluated against a set of metadata elements for each harmonized metadata record in the SWR platform. Records for which harmonisation fails are not evaluated (nor imported).
| Label | Description | Score |
|---|---|---|
| Identification | Unique identification of the dataset (A UUID, URN, or URI, such as DOI) | 10 |
| Title | Short meaningful title | 20 |
| Abstract | Short description or abstract (1/2 page), can include (multiple) scientific/technical references | 20 |
| Author/Organisation | An entity responsible for the resource (e.g. person or organisation) | 20 |
| Date | Last update date | 10 |
| Type | The nature of the content of the resource | 10 |
| Rights | Information about rights and licences | 10 |
| Extent (geographic) | Geographical coverage (e.g. EU, EU & Balkan, France, Wallonia, Berlin) | 5 |
| Extent (temporal) | Temporal coverage | 5 |
Architecture
Technologies Stack
The methodology of ETS/ATS has been suggested to develop validation tests.
Abstract Executable Test Suites (ATS) define a set of abstract test cases or scenarios that describe the expected behaviour of metadata without specifying the implementation details. These test suites focus on the logical aspects of metadata validation and provide a high-level view of metadata validation requirements, enabling stakeholders to understand validation objectives and constraints without getting bogged down in technical details. They serve as a valuable communication and documentation tool, facilitating collaboration between metadata producers, consumers, and validators. ATS are often documented using natural language descriptions, diagrams, or formal specifications. They outline the expected inputs, outputs, and behaviours of the metadata under various conditions.
The SWR ATS is under development at https://github.com/soilwise-he/metadata-validator/blob/main/docs/abstract_test_suite.md
Executable Test Suites (ETS) are sets of tests designed according to ATS to perform the metadata validation. These tests are typically automated and can be run repeatedly to ensure consistent validation results. Executable test suites consist of scripts, programs, or software tools that perform various validation checks on metadata. These checks can include:
- Data Integrity: Checking for inconsistencies or errors within the metadata. This includes identifying missing values, conflicting information, or data that does not align with predefined constraints.
- Standard Compliance: Assessing whether the metadata complies with relevant industry standards, such as Dublin Core, MARC, or specific domain standards like those for scientific data or library cataloguing.
- Interoperability: Evaluating the metadata's ability to interoperate with other systems or datasets. This involves ensuring that metadata elements are mapped correctly to facilitate data exchange and integration across different platforms.
- Versioning and Evolution: Considering the evolution of metadata over time and ensuring that the validation process accommodates versioning requirements. This may involve tracking changes, backward compatibility, and migration strategies.
- Quality Assurance: Assessing the overall quality of the metadata, including its accuracy, consistency, completeness, and relevance to the underlying data or information resources.
- Documentation: Documenting the validation process itself, including any errors encountered, corrective actions taken, and recommendations for improving metadata quality in the future.
ETS is currently implemented through Hale Connect instance and as a locally running prototype of GeoNetwork instance.
Main Sequence Diagram
Metadata INSPIRE compliance
Info
Current version: 0.2.0
Technology: Esdin Test Framework (ETF), Python
Project: Metadata validator
Access point: Postgres database
Compliance to a given standard is an indicator for (meta)data quality. This indicator is measured on datasets claiming to confirm to the INSPIRE regulation. This validation is performed on non augmented, non harmonised metadata records. The observed indicator is stored on the augmentation table. The Esdin Test Framework is used combined with metadata validation rules from the INSPIRE community.
Regarding the INSPIRE validation, all metadata records with the source property value equal to INSPIRE are validated against INSPIRE validation. In total 506 metadata records were harvested from the INSPIRE Geoportal.
For this case, the INSPIRE Reference Validator was used. Validator is based on INSPIRE ATS and is available as a validation service. For the initial validation, INSPIRE metadata were harvested to the local instance of GeoNetwork, which allows on the fly validation of metadata using external validation services (including INSPIRE Reference Validator). Metadata were dowloaded from the PostgreSQL database and uploaded to the local instance of GeoNetwork, where the XML validation and INSPIRE validation were executed. Two validation runs were executed: one to check consistency of metadata using XSD and Schematron (using templates for ISO 19115 standard for spatial data sets and ISO 19119 standard for services), the second for validation of metadata records using INSPIRE ETS.
Architecture
Technologies Stack
| Technology | Description |
|---|---|
| Python | Used for the linkchecker integration, API development, and database interactions. |
| PostgreSQL | Primary database for storing and managing link information. |
| CI/CD | Automated pipeline for continuous integration and deployment, with scheduled weekly runs for link liveliness assessment. |
| Esdin Test Framework | Opensource validation framework, commonly used in INSPIRE. |
Main Sequence Diagram
Database Design
Results of metadata validation are stored on PostgreSQL database, table is called validation in a schema validation.
| identifier | Score | Date |
|---|---|---|
| abc-123-cba | 60 | 2025-01-20T12:20:10Z |
Validation runs every week as a CI-CD pipeline on records which have not been validated for 2 weeks. This builds up a history to understand validation results over time (consider that both changes in records, as well as the ETS itself may cause differences in score).
Integrations & Interfaces
Metadata Augmentation
Introduction
Overview and Scope
This set of components augments metadata statements using various techniques. Augmentations are stored on a dedicated augmentation table, indicating the process which produced it. The statements are combined with the ingested content to offer users an optimal catalogue experience.
At the moment, Metadata Augmentation functionality is covered by the following components:
- Keyword Matcher
- Element Matcher
- Translation Module
- Link Liveliness Assessment
- Spatial Metadata Extractor
- Spatial Locator
- Metadata Interlinker
Upcoming components
Intended Audience
Metadata Augmentation is a backend component providing outputs, which users can see displayed in the Metadata Catalogue. Therefore the Intended Audience corresponds to the one of the Metadata Catalogue. Additionally we expect a maintenance role:
- SWC Administrator monitoring the augmentation processes, access to history, logs and statistics. Administrators can manually start a specific augmentation process.
Keyword matcher
Info
Current version: 0.2.0
Technology: Python
Release: https://doi.org/10.5281/zenodo.14924181
Projects: Keyword matcher
Overview and Scope
Keywords are an important mechanism to filter and cluster records. Similar keywords need to be clustered to be able to match them. This module evaluates keywords of existing records to make them equal in case of high similarity.
Analyses existing keywords on a metadata record. Two cases can be identified:
- If a keyword, having a skos identifier, has a closeMatch or sameAs relation to a prefered keyword, the prefered keyword is used.
- If an existing keyword, without skos identifier, matches a prefered keyword by (translated) string or synonym, then append the matched keyword (including skos identifier).
To facilitate this use case the SWR contains a Knowledge graph of prefered keywords in the soil domain derived from Agrovoc, Gemet and ISO11074. This knowledge graph is maintained at https://github.com/soilwise-he/soil-health-knowledge-graph. These vocabularies are multilingual, facilitating the translation case.
For metadata records which have not been analysed yet (in that iteration), the module extracts the keywords, for each keyword an analyses is made if it matches any of the prefered keywords, the prefered keyword is added to the augmentation results for that record. For string matching a fuzzy match algorithm is used, requiring a 90% match (configurable). Translations are matched using the metadata language as indicated in the record.
Key Features
Architecture
Technological Stack
| Technology | Description |
|---|---|
| Python | Used for the keyword matching and database interactions. |
| PostgreSQL | Primary database for storing and managing information. |
| Docker | Used for containerizing the application, ensuring consistent deployment across environments. |
| * CI/CD | Automated pipeline for continuous integration and deployment, with scheduled dayly runs. |
Main Sequence Diagram
Database Design
Integrations & Interfaces
Key Architectural Decisions
- The process runs as a CI-CD pipeline at daily intervals.
Risks & Limitations
Element Matcher
Overview and Scope
Key Features
Architecture
Technological Stack
Main Sequence Diagram
Database Design
Integrations & Interfaces
Key Architectural Decisions
Risks & Limitations
Translation module
Overview and Scope
Some records arrive in a local language, SWR translates the main properties for the record: title and abstract into English, to offer a single language user experience. The translations are used in filtering and display of records.
The translation module builds on the EU translation service (API documentation at https://language-tools.ec.europa.eu/). Translations are stored in a database for reuse by the SWR.
The EU translation returns asynchronous responses to translation requests, this means that translations may not yet be available after initial load of new data. A callback operation populates the database, from that moment a translation is available to SWR. The translation service uses 2-letter language codes, it means a translation from a 3-letter iso code (as used in for example iso19139:2007) to 2-letter code is required. The EU translation service has a limited set of translations from a certain to alternative language available, else returns an error.
Initial translation is triggered by a running harvester. The translations will then be available once the record is ingested to the triplestore and catalogue database in a followup step of the harvester.
Key Features
Architecture
Technological Stack
| Technology | Description |
|---|---|
| Python | Used for the translation module, API development, and database interactions. |
| PostgreSQL | Primary database for storing and managing information. |
| FastAPI | Employed to create and expose REST API endpoints. Utilizes FastAPI's efficiency and auto-generated Swagger documentation. |
| Docker | Used for containerizing the application, ensuring consistent deployment across environments. |
| CI/CD | Automated pipeline for continuous integration and deployment, with scheduled dayly runs. |
Main Sequence Diagram
Database Design
Integrations & Interfaces
Key Architectural Decisions
Risks & Limitations
Link Liveliness Assessment
Info
Current version: 1.1.4
Technology: Python, FastAPI
Release: https://doi.org/10.5281/zenodo.14923790
Projects: Link liveliness assessment
Overview and Scope
Metadata (and data and knowledge sources) tend to contain links to other resources. Not all of these URIs are persistent, so over time they can degrade. In practice, many non-persistent knowledge sources and assets exist that could be relevant for SWR, e.g. on project websites, in online databases, on the computers of researchers, etc. Links pointing to such assets might however be part of harvested metadata records or data and content that is stored in the SWC.
The Link Liveliness Assessment (LLA) component runs over the available links stored with the SWC assets and checks their status. The function is foreseen to run frequently over the URIs in the SWC, assessing and storing the status of the link.
A link in metadata record either points to:
- another metadata record
- a downloadable instance (pdf/zip/sqlite/mp4/pptx) of the resource
- the resource itself
- documentation about the resource
- identifier of the resource (DOI)
- a webservice or API (sparql, openapi, graphql, ogc-api)
Linkchecker evaluates for a set of metadata records, if:
- the links to external sources are valid
- the links within the repository are valid
- link metadata represents accurately the resource (mime type, size, data model, access constraints)
While evaluating the context of a link, the LLA component may derive some contextual metadata, which can augment the metadata record. These results are stored in the metadata augmentation table. Metadata aspects derived are file size, file format.
Key Features
The LLA component privides the following functions:
- Link validation: Returns HTTP status codes for each link, along with other important information such as the parent URL, any warnings, and the date and time of the test. Additionally, the tool enhances link analysis by identifying various metadata attributes, including file format type (e.g., image/jpeg, application/pdf, text/html), file size (in bytes), and last modification date. This provides users with valuable insights about the resource before accessing it.
- Broken link categorization: Identifies and categorizes broken links based on status codes, including Redirection Errors, Client Errors, and Server Errors.
- Deprecated links identification: Flags links as deprecated if they have failed for X consecutive tests, in our case X equals to 10. Deprecated links are excluded from future tests to optimize performance.
- Timeout management: Allows the identification of URLs that exceed a timeout threshold which can be set manually as a parameter in linkchecker's properties.
- Availability monitoring: When run periodically, the tool builds a history of availability for each URL, enabling users to view the status of links over time.
- OWS services (WMS, WFS, WCS, CSW) typically return a HTTP 500 error when called without the necessary parameters. A handling for these services has been applied in order to detect and include the necessary parameters before being checked.
- On demand URL validation Enables real-time checking of individual URLs without storing results in the database. Returns immediate feedback including status, content metadata, redirect information and diagnostic messages explaining link issues. Particularly useful for pre-validating links before processing in other tools, avoiding unnecessary operations on broken URLs. Utilized in Spatial Metadata Extractor to skip processing broken URLs
A javascript widget is further used to display the link status directly in the SoilWise Metadata Catalogue record.
The API can be used to identify which records have broken links.
Architecture
Technological Stack
| Technology | Description |
|---|---|
| Python | Used for the linkchecker integration, API development, and database interactions. |
| PostgreSQL | Primary database for storing and managing link information. |
| FastAPI | Employed to create and expose REST API endpoints. Utilizes FastAPI's efficiency and auto-generated Swagger documentation. |
| Docker | Used for containerizing the application, ensuring consistent deployment across environments. |
| CI/CD | Automated pipeline for continuous integration and deployment, with scheduled weekly runs for link liveliness assessment. |
Main Component Diagram
flowchart LR
H["Harvester"]-- "writes" -->MR[("Record Table")]
MR-- "reads" -->LAA["Link Liveliness Assessment"]
MR-- "reads" -->CA["Catalogue"]
LAA-- "writes" -->LLAL[("Links Table")]
LAA-- "writes" -->LLAVH[("Validation History Table")]
CA-- "reads" -->API["**API**"]
LLAL-- "writes" -->API
LLAVH-- "writes" -->API
Main Sequence Diagram
sequenceDiagram
participant Linkchecker
participant DB
participant Catalogue
Linkchecker->>DB: Establish Database Connection
Linkchecker->>Catalogue: Extract Relevant URLs
loop URL Processing
Linkchecker->DB: Check URL Existence
Linkchecker->DB: Check Deprecation Status
alt URL Not Deprecated
Linkchecker-->DB: Insert/Update Records
Linkchecker-->DB: Insert/Update Links with file format type, size, last_modified
Linkchecker-->DB: Update Validation History
else URL Deprecated
Linkchecker-->DB: Skip Processing
end
end
Linkchecker->>DB: Close Database Connection
Database Design
classDiagram
Links <|-- Validation_history
Links <|-- Records
Links : +Int ID
Links : +Int fk_records
Links : +String Urlname
Links : +String deprecated
Links : +String link_type
Links : +Int link_size
Links : +DateTime last_modified
Links : +String Consecutive_failures
class Records{
+Int ID
+String Records
}
class Validation_history{
+Int ID
+Int fk_link
+String Statuscode
+String isRedirect
+String Errormessage
+Date Timestamp
}
Integrations & Interfaces
- Visualisation of evaluation in Metadata Catalogue, the assessment report is retrieved using ajax from the each record page
- FastAPI now incorporates additional metadata for links, including file format type, size, and last modified date.
Key Architectural Decisions
Initially we started with linkchecker library, but performance was really slow, because it tested the same links for each page again and again.
We decided to only test the links section of ogc-api:records, it means that links within for example metadata abstract are no longer tested.
OGC OWS services are a substantial portion of links, these services return error 500, if called without parameters. For this scenario we created a dedicated script.
If tests for a resource fail a number of times, the resource is no longer tested, and the resource tagged as deprecated.
Links via a facade, such as DOI, are followed to the page they are referring to. It means the LLA tool can understand the relation between DOI and the page it refers to.
For each link it is known on which record(s) it is mentioned, so if a broken link occurs, we can find a contact to notify in the record.
For the second release we have enhanced the link liveliness assement tool to collect more information about the resources:
- File type format (media type) to help users understand what format they'll be accessing (e.g., image/jpeg, application/pdf, text/html)
- File size to inform users about download expectations
- Last modification date to indicate how recent the resource is
API Updates: The API has been extended to include the newly tracked metadata fields:
- link_type: Shows the file format type of the resource (e.g., image/jpeg, application/pdf)
- link_size: Indicates the size of the resource in bytes
- last_modified: Provides the timestamp when the resource was modified
New Endpoint: - POST /check-url: On-demand URL validation endpoint that accepts a URL and optional check_ogc_capabilities parameter. Returns real-time validation results without database storage, including diagnostic messages to help users understand link issues.
Risks & Limitations
Spatial Metadata Extractor
Overview and Scope
The Spatial Metadata Extractor component identifies and extracts location information from metadata records using multiple approaches:
-
OGC Services Spatial Metadata: Queries OGC services (WMS, WFS, WCS, CSW) if present in metadata links to extract spatial extent and coverage information, providing a robust method for spatial metadata augmentation.
-
Named Entity Recognition (NER): Extracts location entities from metadata titles and abstracts using a trained spaCy model. The component identifies location references in plain text present in the title and abstract, which are then stored as augmentations in the database. This enables spatial discovery and filtering of datasets within the SWC catalogue. The component leverages a trained spaCy model specifically configured to recognize location entities labeled as
Location_positive, ensuring high precision in spatial metadata extraction via the NER approach.
Key Features
The Spatial Metadata Extractor component privides the following functions:
- Detection of OGC service: Automatically identifies if there are URL's present in the metadata that refers an OGC service and, if so, extracts the bounding box from the metadata of this service.
- Location Entity Extraction: Automatically identifies location mentions in metadata titles and abstracts using a trained spaCy NER model.
Architecture
Technological Stack
| Technology | Description |
|---|---|
| Python | Core language for NER pipeline implementation and database interactions. |
| spaCy | Industrial-strength NLP library used for the trained NER model and entity extraction. |
| PostgreSQL | Primary database for storing and managing information. |
| GDAL | Geospatial data abstraction library used for OGC standards augmentation, enabling renabling extraction of bounding box of online geospatial data across formats and services (e.g. WMS, WFS, GML, GeoJSON). |
Main Sequence Diagram
sequenceDiagram
participant Pipeline as Spatial Metadata Extractor Pipeline
participant DB as Database
participant LinkDet as Link Detector
participant OGCCheck as OGC Service Checker
participant OGC as OGC Endpoint (GetCapabilities)
participant GDAL as GDAL
participant Model as trained spaCy Model
Pipeline->>DB: Query unprocessed records
DB-->>Pipeline: Return records (title, abstract, metadata)
loop For each record
%% --- OGC extraction (first) ---
Pipeline->>LinkDet: Detect links in metadata
LinkDet-->>Pipeline: Return candidate URLs
alt URLs found
loop For each URL
Pipeline->>OGCCheck: Check if URL is an OGC service (WMS/WFS/WMTS/WCS/OGC API)
OGCCheck-->>Pipeline: Service type / Not OGC
alt Is OGC service
Pipeline->>OGC: Request GetCapabilities (or equivalent)
OGC-->>Pipeline: Capabilities response (layers + extents)
Pipeline->>GDAL: Parse service metadata and extract bounding box
GDAL-->>Pipeline: Bounding box (minx, miny, maxx, maxy) + CRS
else Not an OGC service
Pipeline-->>Pipeline: Skip OGC extraction for this URL
end
end
else No URLs found
Pipeline-->>Pipeline: Continue without OGC augmentation
end
%% --- NER extraction (second) ---
Pipeline->>Model: Extract locations from title
Model-->>Pipeline: Return location entities
Pipeline->>Model: Extract locations from abstract
Model-->>Pipeline: Return location entities
%% --- persistence ---
Pipeline->>DB: Prepare augmentation rows (OGC bbox + NER entities)
Pipeline->>DB: Prepare status rows
end
Pipeline->>DB: Batch insert augmentations
Pipeline->>DB: Batch insert processing status
DB-->>Pipeline: Confirm insertion
Database Design
The Spatial Metadata Extractor uses the following database structure:
- metadata.records: Source table containing identifier, title, and abstract fields
- metadata.augments: Stores extracted location entities with fields:
record_id: Link to the source recordproperty: Metadata field (e.g., 'title', 'abstract')value: JSON-formatted location data containing text, start_char, and end_charprocess: Set to 'NER-augmentation'
- metadata.augment_status: Tracks processing status with fields:
record_id: Link to the source recordstatus: Processing status (e.g., 'processed')process: Set to 'NER-augmentation'
Example augmentation value:
[
{"text": "Rostock", "start_char": 45, "end_char": 52},
{"text": "Germany", "start_char": 120, "end_char": 127}
]
Integrations & Interfaces
Key Architectural Decisions
- Batch Processing: Records are processed in batches to balance memory usage and database load. Unprocessed records are identified via a LEFT JOIN against the augment_status table.
- Character-Level Offsets: Location entities include start and end character positions to possible enable precise highlighting and linking in the catalogue interface.
- JSON Storage: Extracted locations are stored as JSON arrays to maintain structured data while allowing flexible querying and display options.
- Configurable Model Path: The model path can be specified via environment variable (
MODEL_PATH) or command-line argument, allowing flexibility for different updates of the trained models.
Risks & Limitations
- Model Dependency: Performance and accuracy depend entirely on the quality and training data of the spaCy NER model.
- Entity Label Specificity: the model is trained on a set of entity keywords and then finetuned based on location data already present in the catalogue.
- Abstract And Title Processing: Extraction may fail on malformed abstracts/titles, with errors logged and processing continuing to the next record.
- Database Performance: Large batch operations may impact database performance; batch size may need tuning based on system capacity.
Spatial Locator
Overview and Scope
Key Features
Architecture
Technological Stack
Main Sequence Diagram
Integrations & Interfaces
Key Architectural Decisions
Risks & Limitations
Foreseen functionality
In the next iterations, Metadata augmentation component is foreseen to include the following additional functions:
Keyword extraction
The value of relevant keywords is often underestimated by data producers. This proof-of-concept module evaluates the metadata title/abstract to identify relevant keywords using NLP/NER technology. Integration with the catalogue is foreseen.
Spatial scope analyser
A module that is foreseen to analyse the spatial scope of a resource.
The bounding box will be matched to country or continental bounding boxes using a gazeteer.
To understand if the dataset has a global, continental, national or regional scope
- Retrieves all datasets (as iso19139 xml) from database (records table joined with augmentations) which:
- have a bounding box
- no spatial scope
- in iso19139 format
- For each record it compares the boundingbox to country bounding boxes:
- if bigger then continents > global
- If matches a continent > continental
- if matches a country > national
- if smaller > regional
- result is written to as an augmentation in a dedicated table
EUSO-high-value dataset tagging
The EUSO high-value datasets are those with substantial potential to assess soil health status, as detailed on the EUSO dashboard. This framework includes the concept of soil degradation indicator metadata-based identification and tagging. Each dataset (possibly only those with the supra-national spatial scope - under discussion) will be annotated with a potential soil degradation indicator for which it might be utilised. Users can then filter these datasets according to their specific needs.
The EUSO soil degradation indicators employ specific methodologies and thresholds to determine soil health status, see also the Table below. These methodologies will also be considered, as they may have an impact on the defined thresholds. This issue will be examined in greater detail in the future.
| Soil Degradation | Soil Indicator | Type of methodic for threshold |
|---|---|---|
| Soil erosion | Water erosion | RUSLE2015 |
| Wind erosion | GIS-RWEQ | |
| Tillage erosion | SEDEM | |
| Harvest erosion | Textural index | |
| Post-fire recovery | USLE (Type of RUSLE) | |
| Soil pollution | Arsenic excess | GAMLSS-RF |
| Copper excess | GLM and GPR | |
| Mercury excess | LUCAS topsoil database | |
| Zinc Excess | LUCAS topsoil database | |
| Cadmium Excess | GEMAS | |
| Soil nutrients | Nitrogen surplus | NNB |
| Phosphorus deficiency | LUCAS topsoil database | |
| Phosphorus excess | LUCAS topsoil database | |
| Loss of soil organic carbon | Distance to maximum SOC level | qGAM |
| Loss of soil biodiversity | Potential threat to biological functions | Expert Polling, Questionnaire, Data Collection, Normalization and Analysis |
| Soil compaction | Packing density | Calculation of Packing Density (PD) |
| Salinization | Secondary salinization | - |
| Loss of organic soils | Peatland degradation | - |
| Soil consumption | Soil sealing | Raster remote sense data |
Technically, we forsee the metadata tagging process as illustrated below. At first, metadata record's title, abstract and keywords will be checked for the occurence of specific values from the Soil Indicator and Soil Degradation Codelists, such as Water erosion or Soil erosion (see the Table above). If found, the Soil Degradation Indicator Tag (corresponding value from the Soil Degradation Codelist) will be displayed to indicate suitability of given dataset for soil indicator related analyses. Additionally, a search for corresponding methodology will be conducted to see if the dataset is compliant with the EUSO Soil Health indicators presented in the EUSO Dashboard. If found, the tag EUSO High-value dataset will be added. In later phase we assume search for references to Scientific Methodology papers in metadata record's links. Next, the possibility of involving a more complex search using soil thesauri will also be explored.
flowchart TD
subgraph ic[Indicators Search]
ti([Title Check]) ~~~ ai([Abstract Check])
ai ~~~ ki([Keywords Check])
end
subgraph Codelists
sd ~~~ si
end
subgraph M[Methodologies Search]
tiM([Title Check]) ~~~ aiM([Abstract Check])
kl([Links check]) ~~~ kM([Keywords Check])
end
m[(Metadata Record)] --> ic
m --> M
ic-- + ---M
sd[(Soil Degradation Codelist)] --> ic
si[(Soil Indicator Codelist)] --> ic
em[(EUSO Soil Methodologies list)] --> M
M --> et{{EUSO High-Value Dataset Tag}}
et --> m
ic --> es{{Soil Degradation Indicator Tag}}
es --> m
th[(Thesauri)]-- synonyms ---Codelists
Knowledge Graph
Info
Current version: 0.2.0
Technology: RDF
Project: Soil health knowledge graph
Access point: SWR SPARQL endpoint: https://repository.soilwise-he.eu/sparql
SoilWise develops and implements a knowledge graph linking the knowledge captured in harvested and augmented metadata with various sources of internal and external knowledge sources, particularly taxonomies, vocabularies and ontologies that are also implemented as RDF graphs. Linking such graphs into a harmonized SWR knowledge graph allows reasoning over the relations in the stored graph, and thus allows connecting and smartly combining knowledge from those domains.
The first iteration of the SWR knowledge graph is a graph representation of the (harmonized) metadata that is currently harvested, validated and augmented as part of the SWR catalogue database. It's RDF representation, stored in a triple store, and the SPARQL endpoint deployed on top of the triple store, allow users alternate access to the metadata, exploiting semantics and relations between different assets.
At the same time, preliminary experiments have been conducted to integrate this RDF metadata graph with a dedicated soil health knowledge graph, which is constructed with the assistance of AI/ML, using keyword matching. During this process, unmatched terms from harvested metadata records were identified and cataloged. These terms, acting as candidate keywords, are currently under review by domain experts to assess their potential value for inclusion in the soil health knowledge graph. This analysis serves as a critical mechanism to identify gaps and prioritize new concepts for future enrichment of the graph.
In future iterations, the metadata graph will be linked/merged with this soil health knowledge graph also linking to external resources, establishing a broader interconnected SWR knowledge graph. Consequently, it will evolve into a knowledge network that allows much more powerful and impactful queries and reasoning, e.g. supporting decision support and natural language quering.
Functionality
Knowledge graph querying (SPARQL endpoint)
The SPARQL endpoint, deployed on top of the SWR triple store, allows end users to query the SWR knowledge graph using the SPARQL query language. It is the primary access point to the knowledge graph, both for humans, as well as for machines. Many applications and end users will instead interact with specialised assets that use the SPARQL end-point, such as the Chatbot or the API. However, the SPARQL end-point is the main source for the development of further knowledge applications and provides bespoke search to humans.
Since we are importing resources from various data and knowledge repositories, we expect many duplicities and conflicting statements. Implementation of rules should be permissive, not preventing inclusion, only flag potential inconsistencies.
Ongoing Developments
Knowledge graph enrichment and linking
Info
Access point: https://raw.githubusercontent.com/soilwise-he/soil-health-knowledge-graph/refs/heads/main/soil_health_KG.ttl
Technology: RDF
As a preparation to extend the currently deployed metadata knowledge graph with broader domain knowledge, experimental work has been performed to enrich the knowledge graph to link it with other knowledge graphs.
The following aspects have been worked on and will be further developed and integrated into future iterations of the SoilWise knowledge graph:
- Applying various methods using AI/ML to derive a soil health knowledge graph from unstructured content. This is piloted by using (parts of) the EEA report "Soil monitoring in Europe – Indicators and thresholds for soil health assessments". It tests the effectiveness of various methods to generate knowledge in the form of knowledge graphs from documents, which could also benefit other AI/ML functions foreseen.
- Establishing links between the SoilWise knowledge graph and external taxonomies and ontologies (linked data). Concepts in the SoilWise knowledge graph that (closely) match with concepts in the AGROVOC thesaurus are linked. Other candidate vocabularies in scope are ISO 11074 and GloSIS ontology. The implemented method is exemplary for the foreseen wider linking required to establish a soil health knowledge graph.
- Testing AI/ML based methods to derive additional knowledge (e.g. keywords, geography) for data and knowledge assets. Such methods could for instance be used to further augment metadata or fill exisiting metadata gaps. Besides testing such methods, this includes establishing a model that allows to distinguish between genuine and generated metadata.
Knowledge graph validation
The validation of the soil health knowledge graph will follow a structured two-phase methodology:
Question Formulation
A serie of questions will be developed based on the content of the EEA report. The principle underpinning this step is that if the knowledge graph accurately encapsulates the report’s information, it should generate answers consistent with those derived directly from the source. These questions will undergo collaborative review to ensure their scientific validity and relevance within soil science.
SPARQL Query Execution and Result Verification
Validated questions will be converted into SPARQL queries and executed against the knowledge graph. The retrieved results will be aggregated and systematically cross-referenced with the answers documented in the EEA report. To ensure domain accuracy, a domain expert will perform a rigorous evaluation of the knowledge graph’s outputs, verifying their technical correctness and adherence to soil science principles.
This process ensures the knowledge graph's fidelity to the source material and its capability to support domain-specific queries reliably.
Technology & Integration
Components used:
- Virtuoso (version 07.20.3239)
- Python notebooks
Ontologies/Schemas imported:
- SKOS Core
- Dublin Core
- Agrontology
- QUDT
- PROV-O
- EuroVoc
- Semanticscience Integrated Ontology
- Open Biological and Biomedical Ontology
- Schema.org
- Wikidata
- Biolink
- SWEET ontology
- Allotrope Foundation Ontology
Vocabularies/Thesauri linked:
Data and Knowledge Administration Console
Introduction
Overview & Scope
The Data and Knowledge Administration Console is compiled of several dashboard pages implemented using Apache Superset technology. Apache Superset is an open-source data exploration and visualization platform that enables to create interactive, web-based dashboards with minimal coding. In SoilWise it is used to present statistics from the Metadata Catalogue, results of the Metadata Validation and to provide more insights into the results published by Mission Soil projects. The console is available only for authorised users.
Intended Audience
- SWC Administrator monitoring the contents of the SoilWise Catalogue
- Mission Soil Projects Monitoring officer monitoring the results published by Mission Soil projects
- JRC data analysts monitoring the metadata quality of published soil-related datasets.
Key Features
- Rich visualisation possibilities different kinds of charts, tables and so on.
- Intuitive SQL editor for querying data sources and preparation of data for visualisation.
- WYSIWYG editor for configuring the content and layout of dashboard pages.
- Role-based access control for managing access rights.
Architecture
Technological Stack
| Technology | Description |
|---|---|
| Apache Superset | Used for configuration and display of the dashboard pages compiling the Console. |
| Keycloak | Used for user management and access rigths. |
| PostgreSQL | Connection to a DB with Catalogue contents and validation results. |
Main Components Diagram
User Management and Access Control
User and organisation management, authorisation and authentication are complex, cross-cutting aspects of a system such as the SoilWise repository. Back-end and front-end components need to perform access control for authenticated users. Many organisations already have infrastructures in place, such as an Active Directory or a Single Sign On based on OAuth.
No implementations are yet an integrated part of the SWR delivery, in line with the plan for the first development iteration.
The general model we apply is that:
- a user shall be a member of at least one organisation.
- a user may have at least one role in every organisation that they are a member of.
- a user always acts in the context of one of their roles in one organisation (similar to Github contexts).
- organisations can be hierarchical, and user roles may be inherited from an organisation that is higher up in the hierarchy.
The basic requirements for the SWR authentication mechanisms are:
- User authentication, and thus, provision of authentication tokens, shall be distributed ("Identity Brokering") and may happen through existing services. Authentication mechanisms that are to be supported include OAuth, SAML 2.0 and Active Directory.
- An authoritative Identity Provider, such as an eIDAS-based one, should be integrated in a later iteration as well.
- There shall be a central service that performs role and organisation mapping for authenticated users. This service also provides the ability to configure roles and set up organisations and users. This central service can also provide simple, direct user authentication (username/password-based) for those users who do not have their own authentication infrastructure.
- There may be different levels of trust establishment based on the specific authentication service used. Higher levels of trust may be required to access critical data or infrastructure.
- SWR services shall use Keycloak or JSON Web Tokens for authorization.
- To access SWR APIs, the same rules apply as to access the SWR through the UI.
In later iterations, the authentication and authorisation mechanisms should also be used to facilitate connector-based access to data space resources.
Sign-up
For every registered user of SWR components, an account is needed. This account can be created in one of three ways:
- Automatically, by providing an authentication token that was created by a trusted authentication service and that contains the necessary information on the organisation of the user and the intended role (this can e.g. be implemented through using a DAPS)
- Manually, through self-registration (may only be available for users from certain domains and/or for certain roles)
- Through superuser registration; in this case the user gets issued an activation link and has to set the password to complete registration
Authentication
Certain functionalities of the SWR will be available to anonymous users, but functions that edit any of the state of the system (data, configuration, metadata) require an authenticated user. The easiest form of authentication is to use the login provided by the SWR itself. This log-in is username-password based. A second factor, e.g. through an authenticator app, may be added in the upcoming iteration.
Other forms of authentication include using an existing token.
Authorisation
Every component has to check whether an authenticated user may invoke a desired action based on that user's roles in their organisations. To ensure that the User Interface does not offer actions that a given user may not invoke, the user interface shall also perform authorisation.
Roles are generally defined using Privileges: A certain role may, for example, read certain resources, they may edit or even delete them. Here is an example of such a definition:
A standard user may only read and edit their own User profile, and read the information from their organisation. Once a user has been given the role dataManager, they may perform any CRUD operation on any Data that is in the scope of their organisation. They are also granted read access to publication Theme configurations on their own and in any parent organisations.
Further implementation hints and Technologies
The public cloud hale connect user service can be used for central user management.
Completed work - Iteration 1
- User/Role and Organisation management has been deployed and configured as part of weTransform's hale connect installation.
- As of now, there are three Identity providers deployed as part of that infrastructure:
- The integrated user service in hale connect,
- a Keycloak/OpenID-connect based one using GoPass via Github
- a Data Spaces connector.
Planned work - Iteration 2
- Integrate eIDAS or a different autheoritative Identity Provider
- Update other components to accept the tokens generated by this infrastructure
System & Usage Monitoring
All components and services of the SWR are monitored at different levels to ensure robust operations and security of the system. There will be a central monitoring service for all components that are part of the SWR.
In particular, monitoring needs to fulfill the following requirements:
- For each node, its general state and resource utilisation (RAM, CPU, Volumes) shall be monitored.
- For each container, its general state, e.g. resource consumption (RAM, CPU, Volumes, Transfer, Uptime) shall be monitored.
- For each service, there shall be a health check that can be used to test if the service is responsive and functional, e.g. after a restart.
- If issues that cannot be recovered from automatically occur or which lead to a longer-term degradation of services, messages shall be sent to the operators via channels such as Slack, PagerDuty, or Jira.
- The monitoring system shall provide availability statistics.
- The monitoring system should provide usage statistics.
- The monitoring system may provide a UI element that can be embedded into other components to make usage transparent.
- The monitoring system should provide a dashboard to help system operators with understanding the state of the SWR and to debug incidents, including possible security incidents.
- The monitoring system shall collect warning and error logs to provide guidance for system administrators.
- The monitoring system shall offer the possibility to filter logged interactions based on the https status code, e.g. to identify 404's or 500's.
Technologies
- Grafana
- Portainer
- Prometheus
External integrations
- Jira, Slack, PagerDuty
Usage statistics monitoring
The Soilwise website is using Google Analytics, which uses the first approach.
The SWR catalogue does currently not includes a counter script. The usage can be monitored via the usage logs, which are indexed in an instance of splunk, which provides dashboards on the underlying data.
Future work
Feedback the popularity of resources from usage logs into the catalogue search ranking algorithm.
Interfaces
Introduction
This section focusses on interfaces for access, sharing, population and integration, particularly with EUSO. It describes the technical interfaces (APIs) that will be offered and how they could be exploited for integration. It also describes the user interfaces that are developed as part of SWR. While these are primarily developed to serve the various stakeholder groups of the SoilWise project, these UIs, or derived versions, could in principle also become part of future systems like EUSO.
The architecture of the SoilWise repository (SWR) is highly modular (component based). This allows to compose the infrastructure using a mix of readily available (open source) components, and customized and newly developed elements in a flexible manner. Such a loosely couple, component based architecture allows teams to independently develop, adapt and merge in parts of the architecture without disrupting the working of the system.
For such a system to be flexible and adaptable and to allow agile development, the use of and compliance with standardised interfaces between components, e.g. through APIs, is required. First of all, this makes the system as a whole more flexible and easily adaptable to required changes during the (agile) development process. Second, such interfaces will allow all users (both data & knowledge providers and users) to easily access SWR and/or connect their systems, both for access and usage and for population. Third, it will facilitate the integration of the SWR, or specific parts of SWR with other systems, and particularly with the European Soil Observatory (EUSO). The use of standardised APIs will keep options for such integration open, while both SWR, EUSO, and the required functions they will offer, are evolving.
This section describes the following elements of SWR:
Application Programming Interfaces (APIs)
This page presents an overview of all APIs that are served by the SoilWise Catalogue for (1) internal processing, (2) reuse of SoilWise data and functionality and (3) to enable future integration with other systems, particularly with EUSO. Information on interfaces is also available in a dedicated subchapter per each Technical Component.
Discovery APIs
These APIs allow discovery of (meta)data and knowledge. Most of them are mostly meant to be used as part of SWC backend mechanisms to access or harvest remote data and knowledge resources or to process these resources internally. However, some of them are also relevant for end user discovery of content from SWC. Those user facing discovery APIs are used in user interface components developed by SoilWise, but could also be employed for integration with the EU Soil Observatory and other existing systems that want to make use of SWR.
| Service | Documentation | Purpose |
|---|---|---|
| Catalogue Service for the Web (CSW) | https://repository.soilwise-he.eu/cat/csw | Catalogue service for the web (CSW) is a standardised pattern to interact with (spatial) catalogues, maintained by OGC. |
| OGC API - Records | https://repository.soilwise-he.eu/cat/openapi | OGC is currently in the process of adopting a revised edition of its catalogue standards. The new standard is called OGC API - Records. OGC API - Records is closely related to Spatio Temporal Asset Catalogue (STAC), a community standard in the Earth Observation community. |
| Protocol for metadata harvesting (OAI-PMH) | https://repository.soilwise-he.eu/cat/oaipmh | The open archives initiative has defined a common protocol for metadata harvesting (oai-pmh), which is adopted by many catalogue solutions, such as Zenodo, OpenAire, CKAN. The oai-pmh endpoint of Soilwise can be harvested by these repositories. |
| Spatio Temporal Asset Catalog (STAC) | https://repository.soilwise-he.eu/cat/stac/openapi | TBD |
| OpenSearch | https://repository.soilwise-he.eu/cat/opensearch | TBD |
| SPARQL | https://repository.soilwise-he.eu/sparql/ | The API allows query access to the SoilWise knowledge graph, thus offering querying on linked data, traversing relationships between entities that are relevant and cannot be represented in conventional relational databases. |
| Solr Search API | TBD | The Solr search API Allows query access to the Solr index, so the UI (and other clients) can search the metadata through the index. |
Processing API's
SWC processing APIs are mostly interfaces to components that have been developed or adapted to support the processing of metadata (e.g. metadata augmentation, transforming to RDF) or to support quality assurance and visualisation.
| Service | Documentation | Purpose |
|---|---|---|
| Translate API | https://api.soilwise-he.containers.wur.nl/tolk/docs | This API translates content between languages, and is used for metadata translation. It makes use of the EU translation service https://language-tools.ec.europa.eu/ |
| Link Liveness Assessment API | https://api.soilwise-he.containers.wur.nl/linky/docs | The linkchecker component is designed to evaluate the status, validity and accuracy of links within metadata records in the a OGC API - Records based System. It's responses provide input that is used to inform end users about the status of published links and to collect required data for quality control. |
| RDF to triplestore API | https://repo.soilwise-he.containers.wur.nl/swagger-ui/index.html | Allows the conversion of RDF, e.g. as provided by the CORDIS API's to the SWR triple store. |
| DOI resolution API | TBD | TBD |
| Soil Mission news feed API | TBD | TBD |
User Interfaces (UIs)
The usabilty of the SoilWise Catalogue will eventually not only be determined by it's back-end functionality and the (meta)data and knowledge it exposes. User, whether they are data & knowledge providers, users or SWC administrators, will operate the system through various user interfaces.
Within the first and second development iteration, the following User Interfaces have been deployed as part the SoilWise Catalogue:
Interfaces for end users
SoilWise Metadata Catalogue
Access point: https://repository.soilwise-he.eu/
This UI is currently the main access point for end users searching for resources harvested by SoilWise. It allows users to run term search queries, spatial queries and fulltext search. More information is available in Metadata Catalogue.
Soil Companion
Access point: https://soil-companion.containers.wur.nl/app/index.html
UI providing users the option to query Metadata Catalogue using natural language. Through Chatbot-like user interface, users can ask soil-related question. More information is available in Soil Companion.
Tabular Soil Data Annotation
Access point: https://dataannotator-swr.streamlit.app/
UI providing data publishers support in annotating their tabular data, such as soil observations and measurements. More information is availabl in Tabular Metadata annotation
Soil Vocabulary Viewer
Access point: https://soilwise-he.github.io/soil-vocabs/
UI providing a hierarchical overview of soil properties and corresponding procedures with a link to the related vocabularies. More information is available in Knowledge graph.
Interfaces for administration and system management
Data & Knowledge Administration Console
Access point: https://dashboards.isric.org/superset/dashboard/43/
This UI, based on the superset BI tools, provides a multi-dimensional visual overview of the contents of the SWC together with a dedicated page focusing on Mission Soil outputs. Additionally, results from metadata validations are displayed. More information is available in Data & Knowledge Administration Console.
System Monitoring
Access point: TBD
TBD
Infrastructure
Introduction
This section describes the general hardware infrastructure and deployment pipelines used for the SWR. As of the delivery of this initial version of the technical documentation, a prototype pipeline and hardware environment shall continuously be improved as required to fit the needs of the project.
For the development of First project iteration cycle, we defined the following criteria:
- There is no production environment.
- There is a distributed staging environment, with each partner deploying their solutions to their specific hardware.
- All of the hardware nodes used in the staging environment include an offsite backup capacity, such as a storage box, that is operated in a different physical location.
- There is no central dev/test environment. Each organisation is responsible for its own dev/test environments.
- The deployment and orchestration configuration for this iteration should be stored as YAML in a GitHub repository.
- Deployments to the distributed staging environment are done preferably through GitHub Actions or through alternative pipelines, such as a Jenkins or GitLab instance provided by weTransform or other partners.
- For each component, there shall be separate build processes managed by the responsible partners that result in the built images being made accessible through a hub (e.g. dockerhub)
Work completed - Iteration 1
The Soilwise infrastructure uses components provided by Github. Github components are used to:
- Administer and assign to roles the different Soilwise users.
- Register, prioritise and assign tasks.
- Store source code of software artifacts.
- Author documentation.
- Run CI/CD pipelines.
- Collect user feedback.
During the iteration, the following components have been deployed:
on infrastructure provided by Wageningen University & Research:
- A PostGreSQL database on the PostGreSQL cluster.
- A number of repositories at the WUR Gitlab instance, including CI/CD pipelines to run metadata harvesters.
- A range of services deployed on the WUR k8s cluster, with their configuration stored on Gitlab. Container images are stored on the WUR Harbor repository.
- Usage logs monitored through the WUR instance of Splunk.
- Availability monitoring provided by Uptimerobot.com.
on WeTransform cloud infrastructure:
- a k8s deployment of the Hale Connect stack has been installed and configured. This instance can provide user management and has been integrated with the GitHub repository https://github.com/soilwise-he/Soilwise-credentials. The stack provides Transformation and Validation capabilities.
Future work - Iteration 2
The main objective of iteration 2 is to reorganise the orchestration of the different components, so all components can be centrally accessed and monitored.
The integrations will, whereever feasible, build on API's which are standardised by W3C, OGC or de facto standards, such as Open API or GraphQL.
The intent of the consortium is to set up a distributed architecture, with the staging and production environment in an overall kubernetes-based orchestration mode if it is deemed necessary and advantageous at that point in time.
Containerization
The SWR is being developed in a containerised docker environment. This means that each software component, whether it's a database, storage system, or some kind of service, is compiled into a container image. These images are made available in a hub or repository, so that they can be deployed automatically whenever needed, including to fresh hardware.
GIT versioning system
All aspects of the SoilWise repository can be managed through the SoilWise GitHub repository. This allows all members of the Mission Soil and EUSO community to provide feedback or contribute to any of the aspects.
Documentation
Documentation is maintained in the markdown format using McDocs and deployed as html or pdf using GitHub Pages.
An interactive preview of architecture diagrams is also maintained and published using GitHub Pages.
Source code
Software libraries tailored or developed in the scope of SoilWise are maintained through the GitHub repository.
Container build scripts/deployments
SoilWise is based on an orchestrated set of container deployments. Both the definitions of the containers as well as the orchestration of those containers are maintained through Git.
Harvester definitions
The configuration of the endpoint to be harvested, filters to apply and the interval is stored in a GitHub repository. If the process runs as a CI-CD pipeline, then the logs of each run are also available in Git.
Authored and harvested metadata
Metadata created in SWR, as well as metadata imported from external sources, are stored in GitHub, so a full history of each record is available, and users can suggest changes to existing metadata.
Validation rules
Rules (ATS/ETS) applied to metadata (and data) validation are stored in a git repository.
ETL configuration
Alignments to be applied to the source to be standardised and/or harmonised are stored on a git repository, so users can try the alignment locally or contribute to its development.
Backlog / discussions
Roadmap discussion, backlog and issue management are part of the GitHub repository. Users can flag issues on existing components, documentation or data, which can then be followed up by the participants.
Release management
Releases of the components and infrastructure are managed from a GitHub repository, so users understand the status of a version and can download the packages. The release process is managed in an automated way through CI-CD pipelines.
Glossary
- Abstracting and indexing service (A&I)
- Abstracting and indexing service is a service, e.g. a search engine, that abstracts and indexes documents, and provides matching and ranking functionality in support of information retrieval (source: wikipedia{target=_blank).
- Acceptance Criteria
- Acceptance Criteria can be used to judge if the resulting software satisfies the user's needs. A single user story/requirement can have multiple acceptance criteria.
- API
- Application programming interface (API) is a way for two or more computer programs to communicate with each other (source wikipedia)
- Application profile
- Application profile is a specification for data exchange for applications that fulfil a certain use case. In addition to shared semantics, it also allows for the imposition of additional restrictions, such as the definition of cardinalities or the use of certain code lists (source: purl.eu).
- Artificial Intelligence (AI)
- Artificial Intelligence is a field of study that develops and studies intelligent machines. It includes the fields rule based reasoning, machine learning and natural language processing (NLP). (source: wikipedia)
- Assimilation
- Assimilation is a term indicating the processes involved to combine multiple datasets with different origin into a common dataset, the term is somewhat similarly used in psychology as
incorporation of new concepts into existing schemes(source: wikipedia). But is not well aligned with its usage in the data science community:updating a numerical model with observed data(source: wikipedia) - ATOM
- ATOM is a standardised interface to exchange news feeds over the internet. It has been adopted by INSPIRE as a basic alternative to download services via WFS or WCS.
- Catalogue
- Catalogue or metadata registry is a central location in an organization where metadata definitions are stored and maintained (source: wikipedia)
- Code list
- Code list an enumeration of terms in order to constrain input and avoid errors (source: UN).
- Conceptual model
- Conceptual model or domain model represents concepts (entities) and relationships between them (source: wikipedia)
- Content negotiation
- Content negotiation refers to mechanisms that make it possible to serve different representations of a resource at the same URI (source: wikipedia)
- Controlled vocabulary
- Controlled vocabulary provides a way to organize knowledge for subsequent retrieval. A carefully selected list of words and phrases, which are used to tag units of information so that they are more easily retrieved by a search (source: Semwebtech). Vocabulary, unlike the dictionary and thesaurus, offers an in-depth analysis of a word and its usage in different contexts (source: learn grammar)
- Cordis
- Cordis is the primary source of results from EU-funded projects since 1990
- Corpus
- Corpus (plural: Corpora) is a repository of text documents (knowledge resources); a body of works. Typically the input for information retrieval.
- Catalogue Service for the Web (CSW)
- Catalogue Service for the Web is a standardised protocol from the Open Geospatial Consortium to filter and exchange metadata records. The standard is endorsed by the INSPIRE regulation for discovery services.
- Data
- Data is a collection of discrete or continuous values that convey information, describing the quantity, quality, fact, statistics, other basic units of meaning, or simply sequences of symbols that may be further interpreted formally (Wikipedia).
- Data source
- Data source/provider is a provider of data resources.
- Data management
- Data management is the practice of collecting, organising, managing, and accessing data (for some purpose, such as decision-making).
- Dataset
- Dataset (Also: Data set) A collection of data (Wikipedia).
- Dataverse
- Dataverse is open source research data repository software
- Datacite
- Datacite is a non-profit organisation that provides persistent identifiers (DOIs) for research data.
- Datacite metadata scheme
- Datacite metadata schema a datamodel for metadata for scientific resources
- Destination Earth (DestinE)
- Destination Earth is an initiative of the European Commission to develop a digital model of the Earth to model, monitor and simulate natural phenomena, hazards and the related human activities. The features assist users in designing accurate and actionable adaptation strategies and mitigation measures.
- Digital exchange of soil-related data
- Digital exchange of soil-related data (ISO 28258:2013) presents a conceptual model of a common understanding of what soil profile data are
- Digital soil mapping (DSM)
- Digital soil mapping or pedometric mapping is the creation of soil maps by using field and laboratory observation methods coupled with environmental data through quantitative relationships (source: wikipedia)
- Discovery service
- Discovery service is a concept from INSPIRE indicating a service type which enables discovery of resources (search and find). Typically implemented as CSW.
- Download service
- Download service is a concept from INSPIRE indicating a service type which enables download of a (subset of a) dataset. Typically implemented as WFS, WCS, SOS or Atom.
- Digital Object Identifier (DOI)
- Digital Object Identifier a digital identifier of an object, any object — physical, digital, or abstract
- Encoding
- Encoding is the format used to serialise a resource to a file, common encodings are xml, json, turtle
- Europian soil data centre (ESDAC)
- Europian soil data centre is the thematic centre for soil related data in Europe at the Joint Research Centre
- EU Login
- EU Login is the European Commission's user authentication service. It allows authorised users to access a wide range of Commission web services, using a single email address and password
- European Soil Observatory (EUSO)
- European Soil Observatory is a a dynamic and inclusive platform that aims to support policymaking, by facilitating soil knowledge and data flows, supporting EU Research & Innovation on soils and raising societal awareness of the value of soils
- GDAL OGR
- GDAL and OGR are software packages widely used to interact with a variety of spatial data formats
- Geography Markup Language (GML)
- Geography Markup Language is a standardised encoding for spatial data.
- GeoPackage
- GeoPackage a set of conventions for storing spatial data a SQLite database
- Geoserver
- Geoserver java based software package providing access to remote data through OGC services
- Global Soil Information System (GloSIS)
- Global Soil Information System (GLOSIS) is an activity of FAO Global Soil Partnership enabling a federation of soil information systems and interoperable data sets
- GLOSIS domain model
- GLOSIS domain model is an abstract, architectural component that defines how data are organised; it embodies a common understanding of what soil profile data are.
- GLOSIS Web Ontology
- GLOSIS Web Ontology is an implementation of the GLOSIS domain model using semantic technology
- GLOSIS Codelists
- GLOSIS Codelists is a series of codelists supporting the GLOSIS web ontology. Including the codelists as published in the FAO Guidelines for Soil Description (v2007), soil properties as collected by FAO GfSD and procedures.
- Glosolan
- Glosolan network to strengthen the capacity of laboratories in soil analysis and to respond to the need for harmonizing soil analytical data
- Green Deal
- A program of the European commission setting out a plan to transform Europe’s economy, energy, transport, and industries for a more sustainable future (source: EC).
- Green Deal DataSpace (GDDS)
- Green Deal Dataspace is a German association aiming to build an open ecosystem for resilience and sustainablility, towards the optimization of circular economy and transparency of supply chains.
- Humboldt Alignment Editor (HALE)
- Humboldt Alignment Editor (HALE) java based desktop software to compose and apply a data transformation to data
- Harmonization
- Harmonization is the process of transforming two datasets to a common model, a common projection, usage of common domain values and align their geometries
- Information retreival
- Information retreival (IR) is the task of identifying and retrieving information system resources (e.g. digital objects or metadata records) that are relevant to a search query. It includes searching for the information in a document, searching for the documents themselves, as well as searching for metadata describing the documents.
- Iteration
- An iteration is each development cycle (three foreseen within the SoilWise project) in the project. Each iteration can have phases. There are four phases per iteration focussing on co-design, development, integration and validation, demonstration.
- Joint Research Centre (JRC)
- Joint Research Centre is a part of the European Commission Directorate General. The JRC provides independent, evidence-based science and knowledge, supporting EU policies to positively impact society. Relevant policy areas within JRC are JRC Soil and JRC INSPIRE
- Knowledge
Knowledge is facts, information, and skills acquired through experience or education; the theoretical or practical understanding of a subject. SoilWise mainly considers explicit knowledge -- Information that is easily articulated, codified, stored, and accessed. E.g. via books, web sites, or databases. It does not include implicit knowledge (information transferable via skills) nor tacit knowledge (gained via personal experiences and individual contexts). Explicit knowledge can be further divided into semantic and structural knowledge.
- Semantic knowledge: Also known as declarative knowledge, refers to knowledge about facts, meanings, concepts, and relationships. It is the understanding of the world around us, conveyed through language. Semantic knowledge answers the "What?" question about facts and concepts.
- Structural knowledge: Knowledge about the organisation and interrelationships among pieces of information. It is about understanding how different pieces of information are interconnected. Structural knowledge explains the "How?" and "Why?" regarding the organisation and relationships among facts and concepts.
- Knowledge graph
- Knowledge graph is a representation of a network of real-world entities -- such as objects, events, situations or concepts -- and the relationships between them. Typically the network is made up of nodes, edges, and labels. Both semantic and structural knowledge can be expressed, stored, searched, visualised, and explored as knowledge graphs.
- Knowledge resource
- Knowledge resource is a digital object, such as a document, a web page, or a database, that holds relevant explicit knowledge.
- Knowledge source
- Knowledge source/provider is a provider of knowledge resources.
- Knowledge management
- Knowledge managmenet is the practice of collecting, organising, managing, and accessing knowledge (for some purpose, such as as decision-making).
- Large Language Model (LLM)
- Large Language Model is typically a deep learning model based on the transformer architecture that has been trained on vast amounts of text data, usually from know collections scraped from the Internet.
- Mapserver
- Mapserver C based software package providing access to remote data through OGC services
- Metadata
- (Descriptive) metadata is a summary information describing digital objects such as datasets and knowledge resources.
- Metadata record
- Metadata record is an entry in e.g. a catalogue or abstracting and indexing service with summary information about a digital object.
- Metadata source
- Metadata source/provider is a provider of metadata.
- Mission soil
- Mission Soil is a research and innovation programme of the European Commision, with a strong social science component, putting in place an effective network of 100 living labs and lighthouses to co-create knowledge, test solutions and demonstrate their value in real-life conditions developing a harmonised framework for soil monitoring in Europe, and raising awareness on the vital importance of soils (source: EC research and innovation)
- Mission Soil implementation platform (MIP)
- Mission Implementation Platform is a tool to discover Mission Soil, progress, funded projects, activities and tools to promote cooperation between projects and Mission Soil communities, funding opportunities, news, and events
- Natural Language Processing (NLP)
- Natural Language Processing is an interdisciplinary subfield of computer science and artificial intelligence, primarily concerned with providing computers with the ability to process data encoded in natural language. It is closely related to information retrieval, knowledge representation and computational linguistics.
- Observations, Measurements and Samples (OMS)
- A conceptual model for Observations, Measurements and Samples (OMS), certified as ISO19156:2023
- OGC API
- OGC API building blocks that can be used to assemble novel APIs for web access to geospatial content
- Ontology
- Ontology is a formal representation of the entities in a knowledge graph. Ontologies and knowledge graphs can be expressed in a similar manner and they are closely related. Ontologies can be seen as the (semantic) data model defining classes, relationships and attributes, while knowledge graphs contain the real data according to the (semantic) data model.
- Persistent identifier (PID)
- Persistent identifier is a long-lasting reference to a (digital) object (source wikipedia). In academia, various systems exist which facilitate the creation of PIDs, such as DOI (for publications), ORCID (for authors), and ROR (for organisations).
- Product backlog
- Product backlog is the document where user stories/requirements are gathered with their acceptance criteria, and prioritized.
- QGIS
- QGIS desktop software package to create spatial vizualisations of various types of data
- Retrieval Augmented Generation (RAG)
- Retrieval Augmented Generation is a framework for retrieving facts from an external knowledge base to ground large language models on the most accurate, up-to-date information and enhancing the (pre)trained parameteric (semantic) knowledge with non-parameteric knowledge to avoid hallucinations and get better responses.
- Research Executive Agency (REA)
- The European Research Executive Agency has a mandate is to manage several EU programmes and support services.
- Relational model
- Relational model an approach to managing data using a structure and language consistent with first-order predicate logic (source: wikipedia)
- Resource Description Framework (RDF)
- Resource Description Framework (RDF) a standard model for data interchange on the Web. Common serialisations of RDF are turtle, xml/rdf and json-ld.
- Representational state transfer (REST)
- Representational state transfer a set of guidelines for creating stateless, reliable web APIs (source: wikipedia)
- Reportnet
- Reportnet is the e-Reporting platform for reporting environmental and climate data to the European Environment Agency (EEA). The platform embraces the strategic goals of the European Commission's Green Deal and Digital Strategy and hosts reporting tasks on behalf of EEA and the Commission.
- Requirements
- Requirements are the capabilities of an envisioned component of the repository which are classified as ‘must have’, or ‘nice to have’.
- Rolling plan
- Rolling plan is a methodology for considering the internal and external developments that may generate changes to the SoilWise Repository design and development. It keeps track of any developments and changes on a technical, stakeholder group level or at EUSO/JRC.
- SensorThings API (STA)
- SensorThingsAPI is a formalised protocol to exchange sensor data and tasks between IoT devices, maintained at Open Geospatial Consortium. The protocol can also be used to exchange field and laboratory observations on soils and soil samples.
- Sensor Observation Service (SOS)
- Sensor Observation Service is a formalised protocol to exchange sensor data between entities, maintained at Open Geospatial Consortium.
- Single Sign-On (SSO)
- Single Sign On is an authentication scheme that allows a user to log in with a single ID to any of several related, yet independent, software systems (source: wikipedia).
- Sprint
- Sprint is a small timeframe during which tasks have been defined.
- Sprint backlog
- Sprint backlog is composed of the set of product backlog elements chosen for the sprint, and an action plan for achieving them.
- Soil classification
- Soil classification deals with the systematic categorization of soils based on distinguishing characteristics as well as criteria that dictate choices in use (source: wikipedia)
- Soil health
- Soil health is a state of a soil meeting its range of ecosystem functions as appropriate to its environment. In more colloquial terms, the health of soil arises from favorable interactions of all soil components (living and non-living) that belong together, as in microbiota, plants and animals. It is possible that a soil can be healthy in terms of ecosystem functioning but not necessarily serve crop production or human nutrition directly, hence the scientific debate on terms and measurements.(source: wikipedia)
- Observed property
- An observed property identifies the phenomenon or characteristic that is being measured or observed in a dataset, such as "sea surface height" or "temperature" (source: OGC)
- Soil biological property
- The biological properties refer to the soil biological components in the soil, including microbial life such as bacterial and fungal colonies, which are affected by changes in soil chemical and physical properties, such as acidity and organic matter content (source: Annals of Agricultural Sciences, 2020)
- Soil chemical property
- Soil chemical properties refer to the chemical attributes of soil that influence its ability to supply nutrients to plants, retain harmful elements, and affect plant growth and microbial populations. These properties include nutrient concentrations, pH levels, and the presence of elements such as nitrogen, phosphorus, and potassium, which are critical for soil quality assessment and agricultural productivity. (source: Ecological Indicators, 2022)
- Soil health indicator
- Single characteristic that represents a sustainability effect, whether benefit or negative impact, which may be compared across alternative remediation strategies, comprising one or more remediation (3.380) techniques and/or institutional controls, to evaluate their relative performance (source: iso11074)
- Soil map
- Soil maps are geographical representations showing diversity of soil types or soil properties (soil pH, textures, organic matter, depths of horizons etc.) in an area of interest. It is typically the result of a soil survey inventory, i.e. soil survey. (source: wikipedia)
- Soil physical property
- The physical properties of soil, such as texture, structure, bulk density, porosity, consistency, temperature, colour and resistivity. (source: wikipedia)
- SoilWise Use cases
- The SoilWise use cases are described in the Grant Agreement to understand the needs from the stakeholder groups (users). Each use case provides user stories epics.
- Task
- Task is the smallest segment of work that must be done to complete a user story/requirement.
- Traditional soil mapping
- Traditional soil mapping is the creation of soil maps of spatial distribution of soil properties and soil bodies, by inferring from mental models, leading to manual delineations (source: wikipedia)
- UML
- Unified Modeling Language (UML) a general-purpose modeling language that is intended to provide a standard way to visualize the design of a system (source: wikipedia)
- Usage scenarios
- Usage scenarios describe how (groups of) users might use the software product. These usage scenarios can originate or be updated from the SoilWise use cases, user story epic or user stories/requirements.
- User story
- A User story is a statement, written from the point of view of the user, that describes the functionality needed by the user from the SoilWise Repository.
- User story epic
- A User story epic is a narrative of stakeholders needs that can be narrowed down into smaller specific needs (user stories/requirements).
- Validation framework
- Validation framework is a framework allowing good communication between users and developers, validation of developed products by users, and flexibility on the developer’s side to take change requests into account as soon as possible. The validation framework needs a description of the functionalities to be developed (user stories/requirements), the criteria that enable to verify that the developed component corresponds to the user needs (acceptance criteria), the definition of tasks for the developers (backlog) and the workflow.
- View service
- View service is a concept from INSPIRE indicating a service type which presents a (pre)view of a dataset. Typically implemented as WMS or WMTS.
- Web service
- Web service a service offered by a device to another device, communicating with each other via the Internet (source: wikipedia)
- Web Map service (WMS)
- Web Map service is a formalised protocol to exchange geospatial data represented as images
- Web Feature Service (WFS)
- Web Feature Service is a formalised protocol to exchange geospatial vector data
- Web Coverage Service (WCS)
- Web Coverage Service is a formalised protocol to exchange geospatial grid data
- XML Schema Definition (XSD)
- XML Schema Definition recommendation how to formally describe the elements in an Extensible Markup Language (XML) document (source: wikipedia)